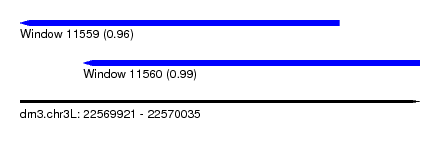
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 22,569,921 – 22,570,035 |
| Length | 114 |
| Max. P | 0.990069 |

| Location | 22,569,921 – 22,570,012 |
|---|---|
| Length | 91 |
| Sequences | 9 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 92.32 |
| Shannon entropy | 0.15854 |
| G+C content | 0.49668 |
| Mean single sequence MFE | -26.68 |
| Consensus MFE | -23.71 |
| Energy contribution | -23.93 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.89 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.72 |
| SVM RNA-class probability | 0.963058 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
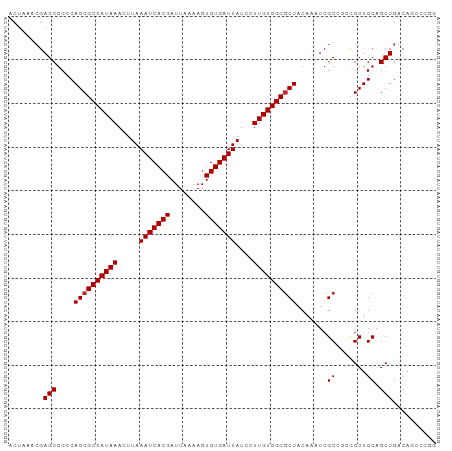
>dm3.chr3L 22569921 91 - 24543557 -------GAGCGCCCAGCGCCAUAAACUUAAAUCACGAUUAAAAGUGUGAUUAUCCUUGUGGCGCUACAAAUCGCCACCGCUGCAGCUGACAGCCCGG -------(.(((...((((((((((.....(((((((........)))))))....))))))))))......))))...(((((....).)))).... ( -27.40, z-score = -2.39, R) >droSim1.chr3L 21862485 98 - 22553184 ACUAAACGAGCGCCCAGCGCCAUAAACUUAAAUCACGAUUAAAAGUGUGAUUAUCCUUGUGGCGCUACAAAUCGCCGCCGCUGCAGCUGACAACCCGG ......((.(((...((((((((((.....(((((((........)))))))....))))))))))......)))))..((....))........... ( -25.50, z-score = -1.54, R) >droSec1.super_11 2496623 98 - 2888827 ACUAAACGAGCGCCCAGCGCCAUAAACUUAAAUCACGAUUAAAAGUGUGAUUAUCCUUGUGGCGCUACAAAUCGCCGCCGCUGCAGCUGACAACCCGG ......((.(((...((((((((((.....(((((((........)))))))....))))))))))......)))))..((....))........... ( -25.50, z-score = -1.54, R) >droYak2.chr3L 23021283 98 - 24197627 ACUAAACGAGCGCCCAGCGCCAUAAACUUAAAUCACGAUUAAAAGUGUGAUUAUCCUUGUGGCGCUACAAAUCGCCGGCGCUGCAGCUGACAACCCGG ........(((...(((((((.........(((((((........)))))))........((((........)))))))))))..))).......... ( -30.00, z-score = -2.45, R) >droEre2.scaffold_4784 22230443 98 - 25762168 ACUAAACGAGCGCCCAGCGCCAUAAACUUAAAUCACGAUUAAAAGUGUGAUUAUCCUUGUGGCGCUACAAAUCGCCGCCGCUGCAGCUGACAACCCGG ......((.(((...((((((((((.....(((((((........)))))))....))))))))))......)))))..((....))........... ( -25.50, z-score = -1.54, R) >droAna3.scaffold_13337 22297714 98 - 23293914 ACUAAACGAGCGCCCAGCGCCAUAAACUUAAAUCACGAUUAAAAGUGUGAUUAUCCUUGUGGCUCUACAAAUCGCGGCUGCUGCAGCUGACAGCUCAG .......((((...(((((((((((.....(((((((........)))))))....)))))))..........((((...)))).))))...)))).. ( -31.20, z-score = -2.98, R) >droVir3.scaffold_13049 8054548 90 + 25233164 ACUAAACGAGCACCCAGCGCCAUAAACUUAAAUCACGAUUAAAAGUGUGAUUAUCCUUGUGGCGCUACAAAUCGC---CGCUGCAGCUGACAG----- ........(((....((((((((((.....(((((((........)))))))....)))))))))).......((---....)).))).....----- ( -25.00, z-score = -2.91, R) >droMoj3.scaffold_6680 2570372 90 + 24764193 ACUAAACGAGCGCCCAGCGCCAUAAACUUAAAUCACGAUUAAAAGUGUGAUUAUCCUUGUGGCGCUACAAAUCGC---CGCUGCAGCUGACAG----- ........(((....((((((((((.....(((((((........)))))))....)))))))))).......((---....)).))).....----- ( -25.00, z-score = -2.31, R) >droGri2.scaffold_15110 15010183 90 - 24565398 ACUAAACGAGCGCCCAGCGCCAUAAACUUAAAUCACGAUUAAAAGUGUGAUUAUCCUUGUGGCGCUACAAAUCGC---CGCUGCAGCUGACAG----- ........(((....((((((((((.....(((((((........)))))))....)))))))))).......((---....)).))).....----- ( -25.00, z-score = -2.31, R) >consensus ACUAAACGAGCGCCCAGCGCCAUAAACUUAAAUCACGAUUAAAAGUGUGAUUAUCCUUGUGGCGCUACAAAUCGCCGCCGCUGCAGCUGACAGCCCGG ........(((((..((((((((((.....(((((((........)))))))....)))))))))).......((....)).)).))).......... (-23.71 = -23.93 + 0.22)

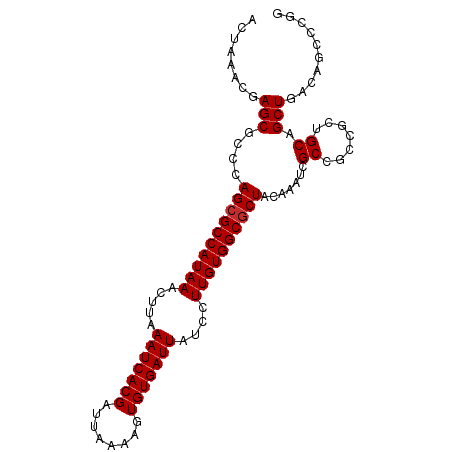
| Location | 22,569,939 – 22,570,035 |
|---|---|
| Length | 96 |
| Sequences | 9 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 84.22 |
| Shannon entropy | 0.31771 |
| G+C content | 0.44656 |
| Mean single sequence MFE | -27.86 |
| Consensus MFE | -22.04 |
| Energy contribution | -22.16 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.79 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.40 |
| SVM RNA-class probability | 0.990069 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 22569939 96 - 24543557 ------------------ACGAGGAACUAGUAACAAGU-----AACGAGCGCCCAGCGCCAUAAACUUAAAUCACGAUUAAAAGUGUGAUUAUCCUUGUGGCGCUACAAAUCGCCACCG ------------------..(((((.........((((-----.....((((...)))).....)))).(((((((........))))))).)))))((((((........)))))).. ( -26.00, z-score = -2.81, R) >droSim1.chr3L 21862503 108 - 22553184 -----------ACGAGGAACUAGUAACAAGUAACAGAAAACUAAACGAGCGCCCAGCGCCAUAAACUUAAAUCACGAUUAAAAGUGUGAUUAUCCUUGUGGCGCUACAAAUCGCCGCCG -----------....((...........(((........)))...((.(((...((((((((((.....(((((((........)))))))....))))))))))......))))))). ( -26.10, z-score = -2.16, R) >droSec1.super_11 2496641 108 - 2888827 -----------ACGAGGAACUAGUAACAAGUAACGGAAAACUAAACGAGCGCCCAGCGCCAUAAACUUAAAUCACGAUUAAAAGUGUGAUUAUCCUUGUGGCGCUACAAAUCGCCGCCG -----------(((((((.........((((..((..........)).((((...)))).....)))).(((((((........))))))).)))))))(((((........).)))). ( -26.40, z-score = -1.69, R) >droYak2.chr3L 23021301 108 - 24197627 -----------ACCGGGAAGUAGUAACAAGUAACAGAAAACUAAACGAGCGCCCAGCGCCAUAAACUUAAAUCACGAUUAAAAGUGUGAUUAUCCUUGUGGCGCUACAAAUCGCCGGCG -----------.((((............(((........)))...(((......((((((((((.....(((((((........)))))))....)))))))))).....))))))).. ( -29.30, z-score = -2.23, R) >droEre2.scaffold_4784 22230461 108 - 25762168 -----------ACGAGGAAGUAGUAACAAGUAACAGAAAACUAAACGAGCGCCCAGCGCCAUAAACUUAAAUCACGAUUAAAAGUGUGAUUAUCCUUGUGGCGCUACAAAUCGCCGCCG -----------....((...........(((........)))...((.(((...((((((((((.....(((((((........)))))))....))))))))))......))))))). ( -26.10, z-score = -1.94, R) >droAna3.scaffold_13337 22297732 108 - 23293914 -----------UGCUGUAACAAGUAGCAAGUAACAGGAAACUAAACGAGCGCCCAGCGCCAUAAACUUAAAUCACGAUUAAAAGUGUGAUUAUCCUUGUGGCUCUACAAAUCGCGGCUG -----------.(((((.....((((..(((.((((((........(.((((...))))).........(((((((........))))))).))).))).))))))).....))))).. ( -27.20, z-score = -1.50, R) >dp4.chrXR_group8 242980 94 - 9212921 ----------------------GUAGCAAGUAACAGGAAACUAAACGAGCACCCAGCGCCAUAAACUUAAAUCACGAUUAAAAGUGUGAUUAUCCUUGUGGCGCUACAAAUCGCCG--- ----------------------...((..((...((....))..))..))....((((((((((.....(((((((........)))))))....))))))))))...........--- ( -26.40, z-score = -3.61, R) >droPer1.super_80 262569 94 + 286535 ----------------------GUAGCAAGUAACAGGAAACUAAACGAGCACCCAGCGCCAUAAACUUAAAUCACGAUUAAAAGUGUGAUUAUCCUUGUGGCGCUACAAAUCGCCG--- ----------------------...((..((...((....))..))..))....((((((((((.....(((((((........)))))))....))))))))))...........--- ( -26.40, z-score = -3.61, R) >droWil1.scaffold_180698 4240286 116 + 11422946 AUGGGGAUGGGGCAAGUACUGUGAGACGGGUAACAAGAAACUAAACGAGCUCCAAGCGCCAUAAACUUAAAUCACGAUUAAAAGUGUGAUUAUCCUUGUGGCGCUACAAAUCGCCG--- ..((.((((((((..((.(((.....)))((........))...))..))))).((((((((((.....(((((((........)))))))....))))))))))....))).)).--- ( -36.80, z-score = -3.09, R) >consensus ___________AC_AGGAACUAGUAACAAGUAACAGGAAACUAAACGAGCGCCCAGCGCCAUAAACUUAAAUCACGAUUAAAAGUGUGAUUAUCCUUGUGGCGCUACAAAUCGCCGCCG .............................................(((......((((((((((.....(((((((........)))))))....)))))))))).....)))...... (-22.04 = -22.16 + 0.11)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:45:13 2011