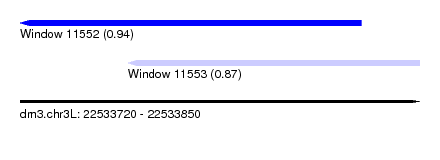
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 22,533,720 – 22,533,850 |
| Length | 130 |
| Max. P | 0.937991 |

| Location | 22,533,720 – 22,533,831 |
|---|---|
| Length | 111 |
| Sequences | 9 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 65.44 |
| Shannon entropy | 0.68834 |
| G+C content | 0.45330 |
| Mean single sequence MFE | -14.50 |
| Consensus MFE | -4.50 |
| Energy contribution | -4.50 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.31 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.42 |
| SVM RNA-class probability | 0.937991 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
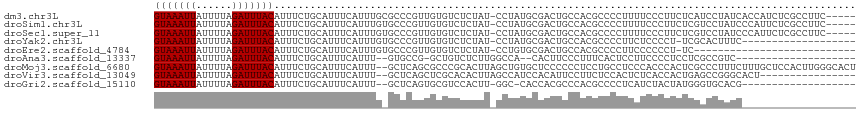
>dm3.chr3L 22533720 111 - 24543557 GUAAAUUAUUUUAGAUUUACAUUUCUGCAUUUCAUUUGCGCCCGUUGUGUCUCUAU-CCUAUGCGACUGCCACGCCCCUUUUCCCUUCUCAUCCUAUCACCAUCUCGCCUUC----- (((((((......)))))))......(((.......)))...(((.(((((.(...-.....).))).)).)))......................................----- ( -12.20, z-score = -1.90, R) >droSim1.chr3L 21833515 111 - 22553184 GUAAAUUAUUUUAGAUUUACAUUUCUGCAUUUCAUUUGUGCCCGUUGUGUCUCUAU-CCUAUGCGACUGCCACGCCCCUUUUCCCUUCUCGUCCUAUCCCAUUCUCGCCUUC----- (((((((......)))))))......((((.......)))).(((.(((((.(...-.....).))).)).)))......................................----- ( -12.40, z-score = -2.21, R) >droSec1.super_11 2468164 111 - 2888827 GUAAAUUAUUUUAGAUUUACAUUUCUGCAUUUCAUUUGUGCCCGUUGUGUCUCUAU-CCUAUGCGACUGCCACGCCCCUUUUCCCUUCUCGUCCUAUCCCAUUCUCGCCUUC----- (((((((......)))))))......((((.......)))).(((.(((((.(...-.....).))).)).)))......................................----- ( -12.40, z-score = -2.21, R) >droYak2.chr3L 22995027 96 - 24197627 GUAAAUUAUUUUAGAUUUACAUUUCUGCAUUUCAUUUGUGCCCGUUGUGUCUCUAU-CCUAUGCGACUGCCACGCCCCUUCUCCCCU-UCGCACUUUC------------------- (((((((......)))))))......((((.......)))).(((.(((((.(...-.....).))).)).))).............-..........------------------- ( -12.40, z-score = -2.09, R) >droEre2.scaffold_4784 22201301 88 - 25762168 GUAAAUUAUUUUAGAUUUACAUUUCUGCAUUUCAUUUGUGCCCGUUGUGUCUCUAU-CCUGUGCGACUGCCACGCCCCUUCCCCCCU-UC--------------------------- (((((((......)))))))......((((.......)))).(((.(((((.(...-.....).))).)).))).............-..--------------------------- ( -12.40, z-score = -2.62, R) >droAna3.scaffold_13337 22266866 92 - 23293914 GUAAAUUAUUUUAGAUUUACAUUUCUGCAUUUCAUUU--GUGCCG-GCUGUCUCUUGGCCA--CACUUCCCUUUCACUCCUUCCCCUCCUCGCCGUC-------------------- (((((((......)))))))......((((.......--)))).(-((((.....))))).--..................................-------------------- ( -13.10, z-score = -2.16, R) >droMoj3.scaffold_6680 2528176 115 + 24764193 GUAAAUUAUUUUAGAUUUACAUUUCUGCAUUUCAUUU--GCUCAGCGCCCGCACUUAGCUGUGCUCCCCCCUCCUGCCUCCCACCCACUCGCCCUUUCUUUGCUCCACUUGGGCACU (((((((......)))))))......(((.......)--))..((.((((((((......))))..........................((.........)).......)))).)) ( -20.90, z-score = -2.17, R) >droVir3.scaffold_13049 8019244 99 + 25233164 GUAAAUUAUUUUAGAUUUACAUUUCUGCAUUUCAUUU--GCUCAGCUCGCACACUUAGCCAUCCACAUUCCUUCUCCACUCUCACCACUGAGCCGGGCACU---------------- (((((((......)))))))................(--((((.(((((.......................................))))).)))))..---------------- ( -14.93, z-score = -2.00, R) >droGri2.scaffold_15110 14973661 94 - 24565398 GUAAAUUAUUUUAGAUUUACAUUUCUGCAUUUCAUUU--GCUCAGUGCGUCCACUU-GGC-CACCACGCCCACGCCCCUCAUCUACUAUGGGUGCACG------------------- (((((((......)))))))......(((.......)--))...((((........-(((-......)))...((((............)))))))).------------------- ( -19.80, z-score = -1.93, R) >consensus GUAAAUUAUUUUAGAUUUACAUUUCUGCAUUUCAUUUG_GCCCGUUGUGUCUCUAU_CCUAUGCGACUGCCACGCCCCUUCUCCCCUCUCGGCCUAUC___________________ (((((((......)))))))................................................................................................. ( -4.50 = -4.50 + 0.00)


| Location | 22,533,755 – 22,533,850 |
|---|---|
| Length | 95 |
| Sequences | 12 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 75.29 |
| Shannon entropy | 0.47068 |
| G+C content | 0.42973 |
| Mean single sequence MFE | -20.43 |
| Consensus MFE | -10.54 |
| Energy contribution | -10.38 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.01 |
| SVM RNA-class probability | 0.872387 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 22533755 95 - 24543557 ---------------------CUGCAGUCAACAAAGGCGCGUAAAUUAUUUUAGAUUUACAUUUCUGCAUUUCAUUUGCG--CCCGUUGUGUCUCUAUCCUAUGCGACUGCCACGCCC- ---------------------..((((((.((((.((.((((((((.....((((........))))......)))))))--))).))))((...........)))))))).......- ( -24.10, z-score = -2.48, R) >droSim1.chr3L 21833550 95 - 22553184 ---------------------CUGCAGUCAACAAAGGCGCGUAAAUUAUUUUAGAUUUACAUUUCUGCAUUUCAUUUGUG--CCCGUUGUGUCUCUAUCCUAUGCGACUGCCACGCCC- ---------------------..((((((.....(((((((((((((......)))))))......((((.......)))--).....))))))(........).)))))).......- ( -21.70, z-score = -1.79, R) >droSec1.super_11 2468199 95 - 2888827 ---------------------CUGCAGUCAACAAAGGCGCGUAAAUUAUUUUAGAUUUACAUUUCUGCAUUUCAUUUGUG--CCCGUUGUGUCUCUAUCCUAUGCGACUGCCACGCCC- ---------------------..((((((.....(((((((((((((......)))))))......((((.......)))--).....))))))(........).)))))).......- ( -21.70, z-score = -1.79, R) >droYak2.chr3L 22995047 95 - 24197627 ---------------------CUGCAGUCAACAAAGGCGCGUAAAUUAUUUUAGAUUUACAUUUCUGCAUUUCAUUUGUG--CCCGUUGUGUCUCUAUCCUAUGCGACUGCCACGCCC- ---------------------..((((((.....(((((((((((((......)))))))......((((.......)))--).....))))))(........).)))))).......- ( -21.70, z-score = -1.79, R) >droEre2.scaffold_4784 22201313 95 - 25762168 ---------------------CUGCAGUCAACAAAGGCGCGUAAAUUAUUUUAGAUUUACAUUUCUGCAUUUCAUUUGUG--CCCGUUGUGUCUCUAUCCUGUGCGACUGCCACGCCC- ---------------------..((((((.(((.(((((((((((((......)))))))......((((.......)))--).....))))))......)))..)))))).......- ( -24.30, z-score = -2.34, R) >droAna3.scaffold_13337 22266883 94 - 23293914 ---------------------CUGCAGUCAACAAAGACGCGUAAAUUAUUUUAGAUUUACAUUUCUGCAUUUCAUUUGUG--CCGGC--UGUCUCUUGGCCACACUUCCCUUUCACUCC ---------------------.(((((((......)))..(((((((......))))))).....))))........(((--..(((--((.....))))).))).............. ( -18.80, z-score = -1.94, R) >dp4.chrXR_group8 210739 96 - 9212921 ---------------------CUGCAGUCAACAAAGACGCGUAAAUUAUUUUAGAUUUACAUUUCUGCAUUUCAUUUGUGCUCUCUUUCUACAGCUAUCUCUUAC-UCUGCCACGCCU- ---------------------..((((.......(((.(((((((((......)))))))......((((.......))))............))..))).....-.)))).......- ( -14.82, z-score = -1.10, R) >droPer1.super_80 230078 96 + 286535 ---------------------CUGCAGUCAACAAAGACGCGUAAAUUAUUUUAGAUUUACAUUUCUGCAUUUCAUUUGUGCUCUCUUUCUACAGCUAUCUCUUAC-UCUGCCACGCCU- ---------------------..((((.......(((.(((((((((......)))))))......((((.......))))............))..))).....-.)))).......- ( -14.82, z-score = -1.10, R) >droWil1.scaffold_180698 4195097 88 + 11422946 ---------------------CUGCAGUCAACAAAGACGCGUAAAUUAUUUUAGAUUUACAUUUCUGCAUUUCAUUUGUG--CAGUGUGCCACACACACACACACACACCC-------- ---------------------.(((((((......)))..(((((((......))))))).....)))).......((((--..(((((.....))))).)))).......-------- ( -19.60, z-score = -2.61, R) >droMoj3.scaffold_6680 2528216 115 + 24764193 CGCGGCAACGCGGCAACGGCAAUGCAGUCAACAAAGACGCGUAAAUUAUUUUAGAUUUACAUUUCUGCAUUUCAUUUGCU--CAGCGC-CCGCACUUAGCUGUGCUCCCCCCUCCUGC- ((((....))))(((..((((((((((((......)))..(((((((......))))))).....))))))......((.--..))))-).((((......))))..........)))- ( -33.20, z-score = -1.64, R) >droVir3.scaffold_13049 8019268 94 + 25233164 ---------------------CUGCAGUCAACAAAGACGCGUAAAUUAUUUUAGAUUUACAUUUCUGCAUUUCAUUUGCU--CAGCUC-GCACACUUAGCCAUCCACAUUCCUUCUCC- ---------------------.(((.(((......)))(((((((((......)))))))......(((.......))).--..))..-)))..........................- ( -14.50, z-score = -2.07, R) >droGri2.scaffold_15110 14973692 82 - 24565398 ---------------------CUGCAGUCAACAAAGAAGCGUAAAUUAUUUUAGAUUUACAUUUCUGCAUUUCAUUUGCU--CAGUGC-GUCCACUUGGCCACCAC------------- ---------------------.(((((((......))...(((((((......)))))))....)))))........(((--.((((.-...)))).)))......------------- ( -15.90, z-score = -1.22, R) >consensus _____________________CUGCAGUCAACAAAGACGCGUAAAUUAUUUUAGAUUUACAUUUCUGCAUUUCAUUUGUG__CCCGUUGUGUCUCUAUCCCAUGCGACUGCCACGCCC_ ......................(((((((......)))..(((((((......))))))).....)))).................................................. (-10.54 = -10.38 + -0.16)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:45:07 2011