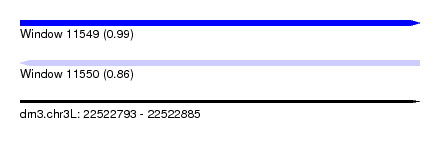
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 22,522,793 – 22,522,885 |
| Length | 92 |
| Max. P | 0.985665 |

| Location | 22,522,793 – 22,522,885 |
|---|---|
| Length | 92 |
| Sequences | 11 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 63.65 |
| Shannon entropy | 0.79781 |
| G+C content | 0.46323 |
| Mean single sequence MFE | -25.90 |
| Consensus MFE | -9.44 |
| Energy contribution | -9.33 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.36 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.21 |
| SVM RNA-class probability | 0.985665 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
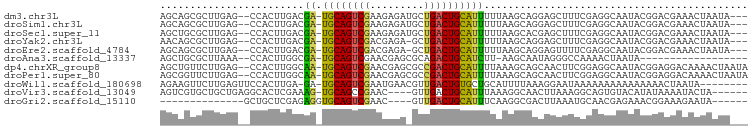
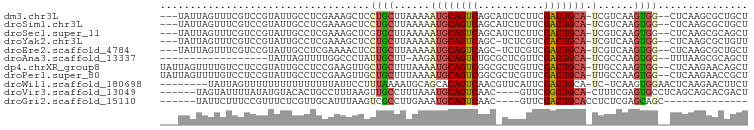
>dm3.chr3L 22522793 92 + 24543557 AGCAGCGCUUGAG--CCACUUGACGA-UGCAGUCGAAGAGAUGCUGACUGCAUUUUUAAGCAGGAGCUUUCGAGGCAAUACGGACGAAACUAAUA--- .((..((...(((--((.(((((.((-((((((((.........)))))))))).)))))...).)))).))..))...................--- ( -26.20, z-score = -1.24, R) >droSim1.chr3L 21824111 92 + 22553184 AGCAGCGCUUGAG--CCACUUGACGA-UGCAGUCGAAGAGAUGCUGACUGCAUUUUUAAGCAGGAGCUUUCGAGGCAAUACGGACGAAACUAAUA--- .((..((...(((--((.(((((.((-((((((((.........)))))))))).)))))...).)))).))..))...................--- ( -26.20, z-score = -1.24, R) >droSec1.super_11 2458937 92 + 2888827 AGCUGCGCUUGAG--CCACUUGACGA-UGCAGUCGAAGAGAUGCUGACUGCAUUUUUAAGCACGAGCUUUCGAGGCAAUACGGACGAAACUAAUA--- .(((.((...(((--((.(((((.((-((((((((.........)))))))))).)))))...).)))).)).)))...................--- ( -25.40, z-score = -1.11, R) >droYak2.chr3L 22985989 91 + 24197627 AACAGCGCUUGAG--CCACUUGACGA-UGCAGUCGACGAGA-GCUGACUGCAUUUUUAAGCAGGAGCUUUCGAGGCAAUACGGACGAAACUAAUA--- ....((.((((((--((.(((((.((-((((((((.(....-).)))))))))).)))))...).))..)))))))...................--- ( -29.20, z-score = -2.94, R) >droEre2.scaffold_4784 22192482 91 + 25762168 AGCAGCGCUUGAG--CCACUUGACGA-UGCAGUCGACGAGA-GCUGACUGCAUUUUUAAGCAGGAGUUUUCGAGGCAAUACGGACGAAACUAAUA--- ....((.((((((--((.(((((.((-((((((((.(....-).)))))))))).)))))..))....))))))))...................--- ( -28.90, z-score = -2.42, R) >droAna3.scaffold_13337 22258953 76 + 23293914 AGCUGCGCUUAAA--CCACUUGGCGA-UGCAGUCGAACGAGCGCAGACUGCAUCUU-AAGCAAUAGGGCCAAAACUAAUA------------------ ......((((((.--((....)).((-(((((((...........)))))))))))-))))..(((........)))...------------------ ( -22.50, z-score = -1.44, R) >dp4.chrXR_group8 200243 95 + 9212921 AGCUGUUCUUGAG--CCACUUGGCAA-UGCAGUCGAACGAGCGCCGACUGCAUUUUAAAGCAGCAACUUCGGAGGCAAUACGGAGGACAAAACUAAUA .((((((.(((((--((....)))((-((((((((.........))))))))))))))))))))..(((((.........)))))............. ( -32.40, z-score = -2.93, R) >droPer1.super_80 220010 95 - 286535 AGCGGUUCUUGAG--CCACUUGGCAA-UGCAGUCGAACGAGCGCCGACUGCAUUUUAAAGCAGCAACUUCGGAGGCAAUACGGAGGACAAAACUAAUA ....((((((..(--((....)))((-((((((((.........))))))))))........((..(....)..))......)))))).......... ( -28.70, z-score = -1.67, R) >droWil1.scaffold_180698 4179953 88 - 11422946 AGAAGUUCUUGAGUUCCACUUGA-GA-UGCAGUCGAAUGAACGUUGACUGUGCUGCAUUUUAAAGGAAUAAAAAAAAAAAAAAACUAAUA-------- ............(((((..((((-((-((((((((......))........)))))))))))).))))).....................-------- ( -16.80, z-score = -1.55, R) >droVir3.scaffold_13049 8003955 87 - 25233164 AGUCGUGCUGCUGAGGCACUCGAAAG-UGCAGCCGAAC----GUUGACUGCAUUUAAAGGCAACUUAAAGGCAGUGUACAUAUAAAAUACUA------ ..(((.(((((((((...))))....-.))))))))..----((..(((((.(((((.(....)))))).)))))..)).............------ ( -23.70, z-score = -0.89, R) >droGri2.scaffold_15110 14956892 74 + 24565398 --------------GCUGCUCGAGAGGUGCAGUCGAAC----GUUGACUGCAUUUCAAGGCGACUUAAAUGCAACGAGAAACGGAAAGAAUA------ --------------.(((((((.(((((((((((((..----.)))))))))))))(((....)))........))))...)))........------ ( -24.90, z-score = -3.02, R) >consensus AGCAGCGCUUGAG__CCACUUGACGA_UGCAGUCGAACAGA_GCUGACUGCAUUUUAAAGCAGGAGCUUUCGAGGCAAUACGGACGAAACUAAUA___ ...........................((((((((.........)))))))).............................................. ( -9.44 = -9.33 + -0.11)
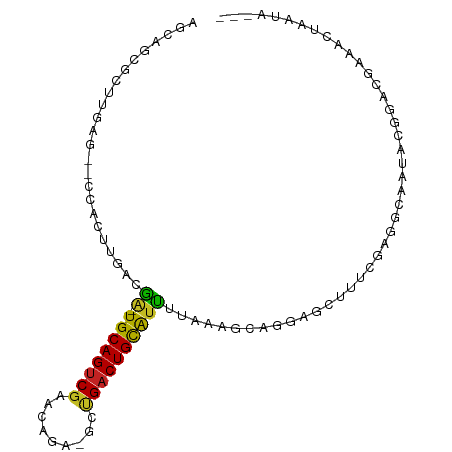
| Location | 22,522,793 – 22,522,885 |
|---|---|
| Length | 92 |
| Sequences | 11 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 63.65 |
| Shannon entropy | 0.79781 |
| G+C content | 0.46323 |
| Mean single sequence MFE | -21.78 |
| Consensus MFE | -8.45 |
| Energy contribution | -8.55 |
| Covariance contribution | 0.10 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.07 |
| Structure conservation index | 0.39 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.97 |
| SVM RNA-class probability | 0.864035 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 22522793 92 - 24543557 ---UAUUAGUUUCGUCCGUAUUGCCUCGAAAGCUCCUGCUUAAAAAUGCAGUCAGCAUCUCUUCGACUGCA-UCGUCAAGUGG--CUCAAGCGCUGCU ---....(((..(((..(....(((.(..((((....))))....((((((((...........)))))))-)......).))--).)..)))..))) ( -22.10, z-score = -0.80, R) >droSim1.chr3L 21824111 92 - 22553184 ---UAUUAGUUUCGUCCGUAUUGCCUCGAAAGCUCCUGCUUAAAAAUGCAGUCAGCAUCUCUUCGACUGCA-UCGUCAAGUGG--CUCAAGCGCUGCU ---....(((..(((..(....(((.(..((((....))))....((((((((...........)))))))-)......).))--).)..)))..))) ( -22.10, z-score = -0.80, R) >droSec1.super_11 2458937 92 - 2888827 ---UAUUAGUUUCGUCCGUAUUGCCUCGAAAGCUCGUGCUUAAAAAUGCAGUCAGCAUCUCUUCGACUGCA-UCGUCAAGUGG--CUCAAGCGCAGCU ---......(((((...(.....)..)))))(((.((((((....((((((((...........)))))))-).(((....))--)..))))))))). ( -25.40, z-score = -1.67, R) >droYak2.chr3L 22985989 91 - 24197627 ---UAUUAGUUUCGUCCGUAUUGCCUCGAAAGCUCCUGCUUAAAAAUGCAGUCAGC-UCUCGUCGACUGCA-UCGUCAAGUGG--CUCAAGCGCUGUU ---.............(((...(((.(..((((....))))....((((((((.((-....)).)))))))-)......).))--)....)))..... ( -23.00, z-score = -1.27, R) >droEre2.scaffold_4784 22192482 91 - 25762168 ---UAUUAGUUUCGUCCGUAUUGCCUCGAAAACUCCUGCUUAAAAAUGCAGUCAGC-UCUCGUCGACUGCA-UCGUCAAGUGG--CUCAAGCGCUGCU ---....(((..(((..(....(((..((....))..((((.(..((((((((.((-....)).)))))))-)..).))))))--).)..)))..))) ( -21.70, z-score = -1.04, R) >droAna3.scaffold_13337 22258953 76 - 23293914 ------------------UAUUAGUUUUGGCCCUAUUGCUU-AAGAUGCAGUCUGCGCUCGUUCGACUGCA-UCGCCAAGUGG--UUUAAGCGCAGCU ------------------..........(((.....(((((-(((((((((((.((....))..)))))))-))(((....))--))))))))..))) ( -24.70, z-score = -1.53, R) >dp4.chrXR_group8 200243 95 - 9212921 UAUUAGUUUUGUCCUCCGUAUUGCCUCCGAAGUUGCUGCUUUAAAAUGCAGUCGGCGCUCGUUCGACUGCA-UUGCCAAGUGG--CUCAAGAACAGCU .....(((((...((.((.........)).))..((..(((...((((((((((((....).)))))))))-))...)))..)--)...))))).... ( -26.70, z-score = -1.20, R) >droPer1.super_80 220010 95 + 286535 UAUUAGUUUUGUCCUCCGUAUUGCCUCCGAAGUUGCUGCUUUAAAAUGCAGUCGGCGCUCGUUCGACUGCA-UUGCCAAGUGG--CUCAAGAACCGCU .....(((((...((.((.........)).))..((..(((...((((((((((((....).)))))))))-))...)))..)--)...))))).... ( -25.80, z-score = -1.08, R) >droWil1.scaffold_180698 4179953 88 + 11422946 --------UAUUAGUUUUUUUUUUUUUUUAUUCCUUUAAAAUGCAGCACAGUCAACGUUCAUUCGACUGCA-UC-UCAAGUGGAACUCAAGAACUUCU --------....(((((((...........((((......(((((((.................).)))))-).-......))))...)))))))... ( -8.69, z-score = 0.60, R) >droVir3.scaffold_13049 8003955 87 + 25233164 ------UAGUAUUUUAUAUGUACACUGCCUUUAAGUUGCCUUUAAAUGCAGUCAAC----GUUCGGCUGCA-CUUUCGAGUGCCUCAGCAGCACGACU ------..((((.......))))(((((.((((((.....)))))).)))))...(----((.(.((((((-((....)))))...))).).)))... ( -20.80, z-score = -0.83, R) >droGri2.scaffold_15110 14956892 74 - 24565398 ------UAUUCUUUCCGUUUCUCGUUGCAUUUAAGUCGCCUUGAAAUGCAGUCAAC----GUUCGACUGCACCUCUCGAGCAGC-------------- ------.................(((((.((((((....)))))).(((((((...----....)))))))........)))))-------------- ( -18.60, z-score = -2.11, R) >consensus ___UAUUAGUUUCGUCCGUAUUGCCUCGAAAGCUCCUGCUUAAAAAUGCAGUCAGC_UCUCUUCGACUGCA_UCGUCAAGUGG__CUCAAGCGCUGCU ..............................................(((((((...........)))))))........................... ( -8.45 = -8.55 + 0.10)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:45:05 2011