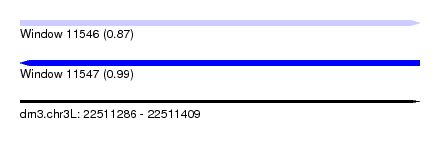
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 22,511,286 – 22,511,409 |
| Length | 123 |
| Max. P | 0.989855 |

| Location | 22,511,286 – 22,511,409 |
|---|---|
| Length | 123 |
| Sequences | 11 |
| Columns | 139 |
| Reading direction | forward |
| Mean pairwise identity | 81.02 |
| Shannon entropy | 0.38686 |
| G+C content | 0.45445 |
| Mean single sequence MFE | -32.43 |
| Consensus MFE | -21.51 |
| Energy contribution | -21.51 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.01 |
| SVM RNA-class probability | 0.873463 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
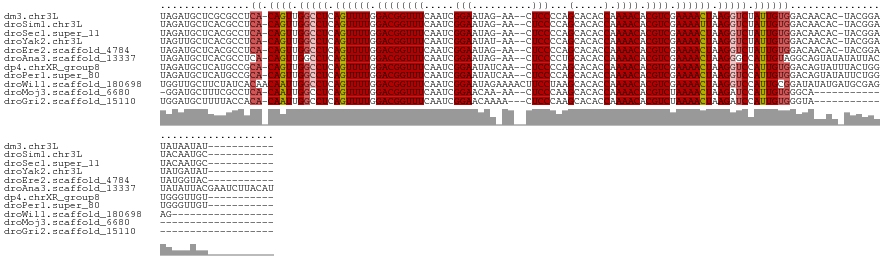
>dm3.chr3L 22511286 123 + 24543557 UAGAUGCUCGCGCCUCA-CAGUUGGCCUCAGUUUUGGACGGUUUCAAUCGGAAUAG-AA--CUCCCCAGCACACCAAAACACGUCGAAAACUAAGGUCUAUUGUGGACAACAC-UACGGAUAUAAUAU----------- (((..((....)).(((-((((.(((((.((((((.((((((((.....(((....-..--.)))...(.....).)))).)))).)))))).))))).)))))))......)-))............----------- ( -29.50, z-score = -0.98, R) >droSim1.chr3L 21812312 123 + 22553184 UAGAUGCUCACGCCUCA-CAGUUGGCCUCAGUUUUGGACGGUUUCAAUCGGAAUAG-AA--CUCCCCAGCACACCAAAACACGUCGAAAAUUAAGGUCUAUUGUGGACAACAC-UACGGAUACAAUGC----------- (((..((....)).(((-((((.(((((.((((((.((((((((.....(((....-..--.)))...(.....).)))).)))).)))))).))))).)))))))......)-))............----------- ( -27.20, z-score = -0.22, R) >droSec1.super_11 2446960 123 + 2888827 UAGAUGCUCACGCCUCA-CAGUUGGCCUCAGUUUUGGACGGUUUCAAUCGGAAUAG-AA--CUCCCCAGCACACCAAAACACGUCGAAAACUAAGGUCUAUUGUGGACAACAC-UACGGAUACAAUGC----------- (((..((....)).(((-((((.(((((.((((((.((((((((.....(((....-..--.)))...(.....).)))).)))).)))))).))))).)))))))......)-))............----------- ( -29.50, z-score = -0.85, R) >droYak2.chr3L 22973857 123 + 24197627 UAGUUGCUCACGCCUCA-CAGUUGGCCUCAGUUUUGGACGGUUUCAAUCGGAAUAU-AA--CUCCCCAGCACACCAAAACACGUCGAAAACUAAGGUCUAUUGUGGACAACAC-UACGGAUAUGAUAU----------- ..((((.((......((-((((.(((((.((((((.((((((((.....(((....-..--.)))...(.....).)))).)))).)))))).))))).)))))))))))).(-....).........----------- ( -31.00, z-score = -1.50, R) >droEre2.scaffold_4784 22180743 123 + 25762168 UAGAUGCUCACGCCUCA-CAGUUGGCCUCAGUUUUGGACGGUUUCAAUCGGAAUAG-AA--CUCCCCAGCACACCAAAACACGUCGAAAACUAAGGUCUAUUGUGGACAACAC-UACGGAUAUGGUAC----------- ...........((((((-((((.(((((.((((((.((((((((.....(((....-..--.)))...(.....).)))).)))).)))))).))))).)))))))......(-....)....)))..----------- ( -32.30, z-score = -1.21, R) >droAna3.scaffold_13337 22247675 135 + 23293914 UAGAUGCUCACGCCUCA-CAGUUGGCCUCAGUUUUGGACGGUUUCAAUCGGAAUAG-AA--CUCCCCUGCACACCAAAACACGUCGAAAACUAAGGGCCAUUGUAGGCAGUAUAUAUUACUAUAUUACGAAUCUUACAU .((((..((..((((.(-((((.(((((.((((((.((((((((((...(((....-..--.)))..)).......)))).)))).))))))..))))))))))))))((((.....)))).......))))))..... ( -37.01, z-score = -2.95, R) >dp4.chrXR_group8 186475 125 + 9212921 UAGAUGCUCAUGCCGCA-CAGUUGGCCUCAGUUUUGGACGGUUUCAAUCGGAAUAUCAA--CUCCCCAGCACACCAAAACACGUCGAAAACUAAGGUCCAUUGUGGACAGUAUUUACUGGUGGGUUGU----------- .....((((((....((-((((.(((((.((((((.((((((((.....(((.......--.)))...(.....).)))).)))).)))))).))))).))))))..((((....))))))))))...----------- ( -37.50, z-score = -1.40, R) >droPer1.super_80 206044 125 - 286535 UAGAUGCUCAUGCCGCA-CAGUUGGCCUCAGUUUUGGACGGUUUCAAUCGGAAUAUCAA--CUCCCCAGCACACCAAAACACGUCGAAAACUAAGGUCCAUUGUGGACAGUAUAUUCUGGUGGGUUGU----------- .....((((((....((-((((.(((((.((((((.((((((((.....(((.......--.)))...(.....).)))).)))).)))))).))))).))))))..(((......)))))))))...----------- ( -36.50, z-score = -1.19, R) >droWil1.scaffold_180698 4160346 122 - 11422946 UGGUUGCUUCUAUCACAACAAUUGGCCUCAGUUUUGGACGGUUUCAAUCGGAAUAGAAAACUUCCUAAGCACACCAAAACACGUCGAAAACUAAGGUCCAUUGCGGAUAUAUGAUGCGAGAG----------------- ...((((.((((((....((((.(((((.((((((.((((((((......(..(((........)))..)......)))).)))).)))))).))))).))))..))))...)).))))...----------------- ( -32.60, z-score = -2.18, R) >droMoj3.scaffold_6680 2490401 104 - 24764193 -GGAUGCUUUCGCCUCA-CAAUUGGCCUCAGUUUUGGACGGUUUCAAUCGGAACAA-AA--CUCCCAAGCACACCAAAACACGUCUAAAACUAAGAUCCAUUGUGGGCA------------------------------ -..........(((.((-((((.(..((.((((((((((((((((....)))))..-..--((....))............))))))))))).))..).))))))))).------------------------------ ( -32.90, z-score = -3.69, R) >droGri2.scaffold_15110 14942019 105 + 24565398 UGGAUGCUUUUACCACA-CAAUUGGCCUCAGUUUUGGACGGUUUCAAUCGGAACAAAA---CUCCCAAGCACACCAAAACACGUCUAAAACUAAGAUCCAUUGUGGGUA------------------------------ ..........((((.((-((((.(..((.((((((((((((((((....)))))....---((....))............))))))))))).))..).))))))))))------------------------------ ( -30.70, z-score = -3.02, R) >consensus UAGAUGCUCACGCCUCA_CAGUUGGCCUCAGUUUUGGACGGUUUCAAUCGGAAUAG_AA__CUCCCCAGCACACCAAAACACGUCGAAAACUAAGGUCCAUUGUGGACAACAC_UACGGAUAUAAUAC___________ ............((....((((.(((((.((((((.((((((((.....(((..........)))...(.....).)))).)))).)))))).))))).)))).))................................. (-21.51 = -21.51 + 0.00)
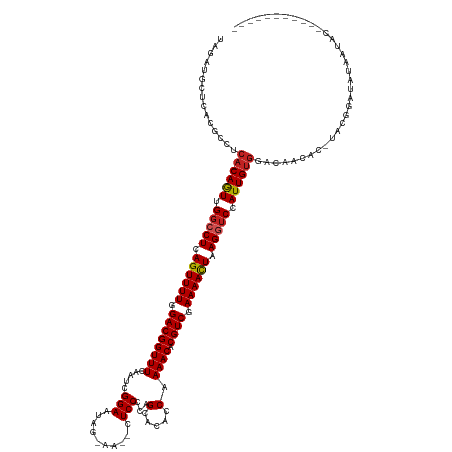
| Location | 22,511,286 – 22,511,409 |
|---|---|
| Length | 123 |
| Sequences | 11 |
| Columns | 139 |
| Reading direction | reverse |
| Mean pairwise identity | 81.02 |
| Shannon entropy | 0.38686 |
| G+C content | 0.45445 |
| Mean single sequence MFE | -37.83 |
| Consensus MFE | -29.33 |
| Energy contribution | -28.22 |
| Covariance contribution | -1.11 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.78 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.39 |
| SVM RNA-class probability | 0.989855 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 22511286 123 - 24543557 -----------AUAUUAUAUCCGUA-GUGUUGUCCACAAUAGACCUUAGUUUUCGACGUGUUUUGGUGUGCUGGGGAG--UU-CUAUUCCGAUUGAAACCGUCCAAAACUGAGGCCAACUG-UGAGGCGCGAGCAUCUA -----------...........((.-(((((...((((...(.((((((((((.((((.((((..((......((((.--..-...)))).))..)))))))).)))))))))).)...))-)).)))))..))..... ( -35.40, z-score = -1.27, R) >droSim1.chr3L 21812312 123 - 22553184 -----------GCAUUGUAUCCGUA-GUGUUGUCCACAAUAGACCUUAAUUUUCGACGUGUUUUGGUGUGCUGGGGAG--UU-CUAUUCCGAUUGAAACCGUCCAAAACUGAGGCCAACUG-UGAGGCGUGAGCAUCUA -----------((((((......))-))))....((((...(.(((((.((((.((((.((((..((......((((.--..-...)))).))..)))))))).)))).))))).)...))-))..((....))..... ( -30.40, z-score = 0.35, R) >droSec1.super_11 2446960 123 - 2888827 -----------GCAUUGUAUCCGUA-GUGUUGUCCACAAUAGACCUUAGUUUUCGACGUGUUUUGGUGUGCUGGGGAG--UU-CUAUUCCGAUUGAAACCGUCCAAAACUGAGGCCAACUG-UGAGGCGUGAGCAUCUA -----------((((((......))-))))....((((...(.((((((((((.((((.((((..((......((((.--..-...)))).))..)))))))).)))))))))).)...))-))..((....))..... ( -36.40, z-score = -1.19, R) >droYak2.chr3L 22973857 123 - 24197627 -----------AUAUCAUAUCCGUA-GUGUUGUCCACAAUAGACCUUAGUUUUCGACGUGUUUUGGUGUGCUGGGGAG--UU-AUAUUCCGAUUGAAACCGUCCAAAACUGAGGCCAACUG-UGAGGCGUGAGCAACUA -----------..............-..((((..(((.......(((((((((.((((.((((..((......((((.--..-...)))).))..)))))))).)))))))))(((.....-...))))))..)))).. ( -37.50, z-score = -1.97, R) >droEre2.scaffold_4784 22180743 123 - 25762168 -----------GUACCAUAUCCGUA-GUGUUGUCCACAAUAGACCUUAGUUUUCGACGUGUUUUGGUGUGCUGGGGAG--UU-CUAUUCCGAUUGAAACCGUCCAAAACUGAGGCCAACUG-UGAGGCGUGAGCAUCUA -----------..............-((..((((((((...(.((((((((((.((((.((((..((......((((.--..-...)))).))..)))))))).)))))))))).)...))-)).))))...))..... ( -35.50, z-score = -1.02, R) >droAna3.scaffold_13337 22247675 135 - 23293914 AUGUAAGAUUCGUAAUAUAGUAAUAUAUACUGCCUACAAUGGCCCUUAGUUUUCGACGUGUUUUGGUGUGCAGGGGAG--UU-CUAUUCCGAUUGAAACCGUCCAAAACUGAGGCCAACUG-UGAGGCGUGAGCAUCUA .....((((.((((.((((....))))))).(((((((.(((((.((((((((.((((.((((..((......((((.--..-...)))).))..)))))))).)))))))))))))..))-).))))....).)))). ( -43.70, z-score = -2.99, R) >dp4.chrXR_group8 186475 125 - 9212921 -----------ACAACCCACCAGUAAAUACUGUCCACAAUGGACCUUAGUUUUCGACGUGUUUUGGUGUGCUGGGGAG--UUGAUAUUCCGAUUGAAACCGUCCAAAACUGAGGCCAACUG-UGCGGCAUGAGCAUCUA -----------...................((((((((.(((.((((((((((.((((.((((..((.......((((--(....))))).))..)))))))).)))))))))))))..))-)).)))).......... ( -38.80, z-score = -1.69, R) >droPer1.super_80 206044 125 + 286535 -----------ACAACCCACCAGAAUAUACUGUCCACAAUGGACCUUAGUUUUCGACGUGUUUUGGUGUGCUGGGGAG--UUGAUAUUCCGAUUGAAACCGUCCAAAACUGAGGCCAACUG-UGCGGCAUGAGCAUCUA -----------..........(((......((((((((.(((.((((((((((.((((.((((..((.......((((--(....))))).))..)))))))).)))))))))))))..))-)).))))......))). ( -39.50, z-score = -2.02, R) >droWil1.scaffold_180698 4160346 122 + 11422946 -----------------CUCUCGCAUCAUAUAUCCGCAAUGGACCUUAGUUUUCGACGUGUUUUGGUGUGCUUAGGAAGUUUUCUAUUCCGAUUGAAACCGUCCAAAACUGAGGCCAAUUGUUGUGAUAGAAGCAACCA -----------------.....((.((...((((((((((((.((((((((((.((((.((((..((..(..(((((....)))))..)..))..)))))))).)))))))))))).)))).)).)))))).))..... ( -39.20, z-score = -3.84, R) >droMoj3.scaffold_6680 2490401 104 + 24764193 ------------------------------UGCCCACAAUGGAUCUUAGUUUUAGACGUGUUUUGGUGUGCUUGGGAG--UU-UUGUUCCGAUUGAAACCGUCCAAAACUGAGGCCAAUUG-UGAGGCGAAAGCAUCC- ------------------------------.(((((((((((.((((((((((.((((.((((..((.....(((((.--..-...)))))))..)))))))).)))))))))))).))))-)).)))..........- ( -40.50, z-score = -4.59, R) >droGri2.scaffold_15110 14942019 105 - 24565398 ------------------------------UACCCACAAUGGAUCUUAGUUUUAGACGUGUUUUGGUGUGCUUGGGAG---UUUUGUUCCGAUUGAAACCGUCCAAAACUGAGGCCAAUUG-UGUGGUAAAAGCAUCCA ------------------------------((((((((((((.((((((((((.((((.((((..((.....(((((.---.....)))))))..)))))))).)))))))))))).))))-)).)))).......... ( -39.20, z-score = -4.36, R) >consensus ___________ACAUCAUAUCCGUA_GUGUUGUCCACAAUGGACCUUAGUUUUCGACGUGUUUUGGUGUGCUGGGGAG__UU_CUAUUCCGAUUGAAACCGUCCAAAACUGAGGCCAACUG_UGAGGCGUGAGCAUCUA ..............................((((((((...(.((((((((((.((((.((((..((.......((((........)))).))..)))))))).)))))))))).)...)).)).)))).......... (-29.33 = -28.22 + -1.11)
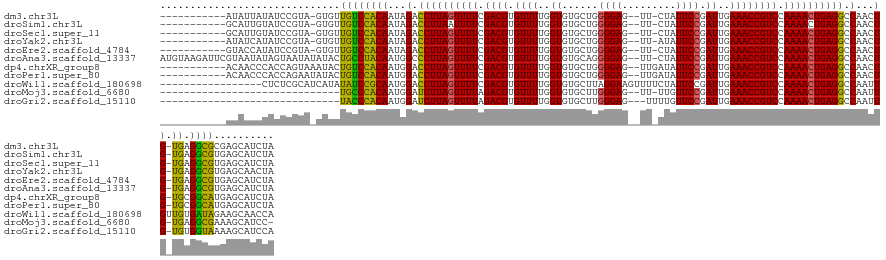

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:45:02 2011