
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 6,145,799 – 6,145,892 |
| Length | 93 |
| Max. P | 0.513284 |

| Location | 6,145,799 – 6,145,892 |
|---|---|
| Length | 93 |
| Sequences | 8 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 63.66 |
| Shannon entropy | 0.71777 |
| G+C content | 0.40298 |
| Mean single sequence MFE | -12.77 |
| Consensus MFE | -5.34 |
| Energy contribution | -5.59 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -0.71 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.513284 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 6145799 93 - 23011544 UAAUUGCUAAAAGGUAUUUAAUUCGUGAUGCUUUUAUAUUUU-----GCCGAUUU-CACUUUGG-GCCCCCCUCAUCGCCACUCCACCCACUCAAUAUUC .....((..(((((((((........))))))))).......-----(((.(...-.....).)-))..........))..................... ( -11.20, z-score = 0.31, R) >droYak2.chr2L 15557088 84 + 22324452 UAAUUGCUAAAAGGUAUUUAAUUCGUGAUGCUUUUAUAUUUC-----GCCGAUUU-CACUUUGAUGCCCCCACCA--CCCACUCCACCGAAA-------- .....((...(((((....((((.((((............))-----)).)))).-.)))))...))........--...............-------- ( -8.30, z-score = 0.01, R) >droEre2.scaffold_4929 15071927 70 - 26641161 UAAUUGCUAAAAGGUAUUUAAUUCGUGAUGCUUUUAUAUUUC-----GCCGAUUU-CACUUUG--GCCCCCAUCA--GCC-------------------- .....(((.(((((((((........))))))))).......-----(((((...-....)))--)).......)--)).-------------------- ( -14.00, z-score = -1.75, R) >droGri2.scaffold_15252 14472628 86 + 17193109 UAAUUGCUAAAAGGUAUUUAAUUCGUGAUGCUUUUAUAUUUCAGCG-UUUAUCCU-CAUUUCAUUCUACGCUCUACA--CAAGCUUCUCG---------- .....(((.(((((((((........))))))))).......((((-(.......-...........))))).....--..)))......---------- ( -12.37, z-score = -1.72, R) >droMoj3.scaffold_6500 18890999 90 + 32352404 UAAUUGCUAAAAGGUAUUUAAUUCGCGUCGCUUUUAGAUUUCAGCGCUUUAUUCUCCAUUUCAUUCAAUGCUCGGCGUUUCAGCUACUCG---------- .....(((....((....(((..((((((.......)))....)))..)))....)).........(((((...)))))..)))......---------- ( -10.70, z-score = 1.15, R) >droAna3.scaffold_12916 8051832 94 - 16180835 UAAUUGCUAAAAGGUAUUUAAUUCCUGAUGCUUUUAUAUUUC-----AUUCGGCU-CACUUUACCCUCCAGACCGCCCAACCUCCGCCCGCUGAGAAGUC .....(((.(((((((((........)))))))))...((((-----(...(((.-.............................)))...)))))))). ( -11.11, z-score = 0.85, R) >dp4.chr4_group1 5262681 82 + 5278887 UAAUUGCUAAAAGGUAUUUAAUUCGUGAUGCUUUUAUAUUUUCAGCGAGAGAGCU-CACUUUAC-UCUCUCCUCGCCCCCCCUU---------------- .....((..(((((((((........)))))))))........((.(((((((..-.......)-)))))))).))........---------------- ( -16.90, z-score = -2.25, R) >droPer1.super_1 10267201 89 + 10282868 UAAUUGCUAAAAGGUAUUUAAUUCGUGAUGCUUUUAUAUUUUCAGCGAGAGAGCU-CACUUUAC-UCUCUCCUCGCCUCCUCGCCCCCCCC--------- .....((..(((((((((........))))))))).........(((((((((..-........-.)))).)))))......)).......--------- ( -17.60, z-score = -2.25, R) >consensus UAAUUGCUAAAAGGUAUUUAAUUCGUGAUGCUUUUAUAUUUC_____ACCGAUCU_CACUUUAC_CCCCCCACCACCCCCACUCCACCCG__________ .........(((((((((........)))))))))................................................................. ( -5.34 = -5.59 + 0.25)
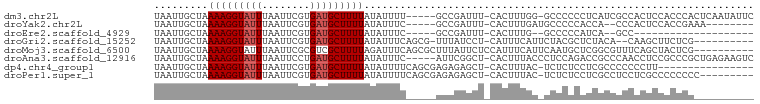

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:20:30 2011