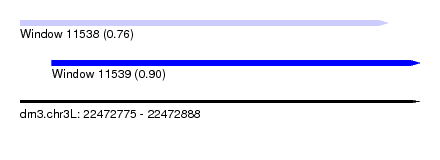
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 22,472,775 – 22,472,888 |
| Length | 113 |
| Max. P | 0.900673 |

| Location | 22,472,775 – 22,472,879 |
|---|---|
| Length | 104 |
| Sequences | 12 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 61.11 |
| Shannon entropy | 0.83934 |
| G+C content | 0.50048 |
| Mean single sequence MFE | -31.84 |
| Consensus MFE | -11.14 |
| Energy contribution | -11.16 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.02 |
| Structure conservation index | 0.35 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.755172 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
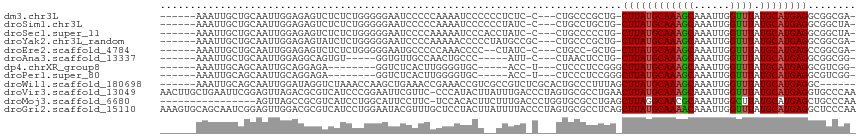
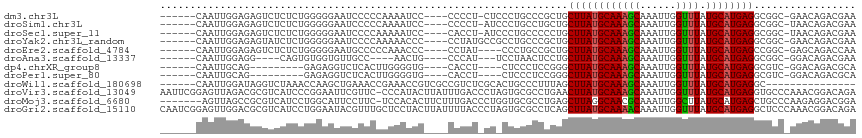
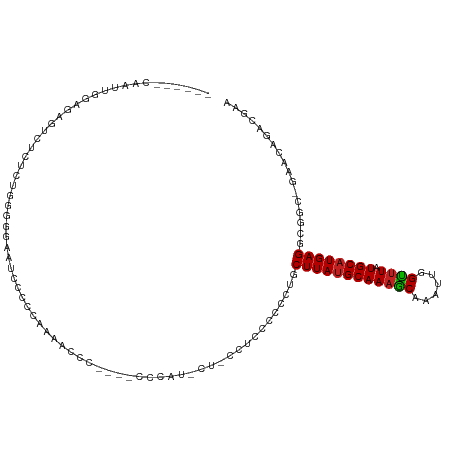
>dm3.chr3L 22472775 104 + 24543557 ------AAAUUGCUGCAAUUGGAGAGUCUCUCUGGGGGAAUCCCCCAAAAUCCCCCCUCUC-C---CUGCCCGCUG-CUUAUGCAAAGCAAAUUGGUUUAUGCAUGAGGCGGCGA- ------........(((...((((((.......((((((...........)))))))))))-)---.))).(((((-((((((((((((......)))).))))))).)))))).- ( -40.41, z-score = -2.89, R) >droSim1.chr3L 21782557 104 + 22553184 ------AAAUUGCUGCAAUUGGAGAGUCUCUCUGGGGGAAUCCCCCAAAAUCCCCCCUAUC-C---CUGCCUGCUG-CUUAUGCAAAGCAAAUUGGUUUAUGCAUGAGGCGGCUA- ------........(((...((((.....))))((((((...........)))))).....-.---.)))..((((-((((((((((((......)))).))))))).)))))..- ( -33.60, z-score = -1.04, R) >droSec1.super_11 2408578 104 + 2888827 ------AAAUUGCUGCAAUUGGAGAGUCUCUCUGGGGGAAUCCCCAAAAAUCCCACCUAUC-C---CUGCCCCCUG-CUUAUGCAAAGCAAAUUGGUUUAUGCAUGAGGCGGCUA- ------.....(((((....((((.....))))(((((.......................-.---...)))))..-((((((((((((......)))).)))))))))))))..- ( -31.53, z-score = -0.77, R) >droYak2.chr3L_random 99245 105 + 4797643 ------AAAUUGCUGCAAUUGGAGAGUAUCUCUGGGGGAAUCCCCAAAAACCCCCUAUGCCGC---CUGCCCGCUG-CUUAUGCAAAGCAAAUUGGUUUAUGCAUGAGGCGGCGA- ------.....((.(((...((((.....))))(((((............)))))..))).))---.....(((((-((((((((((((......)))).))))))).)))))).- ( -38.10, z-score = -1.86, R) >droEre2.scaffold_4784 22142233 101 + 25762168 ------AAAUUGCUGCAAUUGGAGAGUCUCUCUGGGGGAAUGCCCCCAAACCCC--CUAUC-C---CUGCC-GCUG-CUUAUGCAAAGCAAAUUGGUUUAUGCAUGAGCCGGCGA- ------........(((...(((.((......((((((....))))))......--)).))-)---.)))(-((((-((((((((((((......)))).)))))))).))))).- ( -37.10, z-score = -2.35, R) >droAna3.scaffold_13337 22210632 94 + 23293914 ------AAAUUGCUGCAAUUGGAGGCAGUGU-----GGUGUUGCCAACUGCCC-----AUU-C---CUAACUCCUG-CUUAUGCAAAGCAAAUUGGUUUAUGCAUGAGGCGGCGG- ------....((((((....(((((((((.(-----((.....))))))))).-----..)-)---).........-((((((((((((......)))).)))))))))))))).- ( -36.40, z-score = -2.40, R) >dp4.chrXR_group8 140310 92 + 9212921 ------AAAUUGCAGCAAUUGCAGGAGA--------GGUCUCACUUGGGGUGC-----ACC-U---CUCCCUCCGGGCUUAUGCAAAGCAAAUUGGUUUAUGCAUGAGGCGUCGG- ------....(((((...)))))(((((--------(((..(((.....))).-----)))-)---))))..(((((((((((((((((......)))).))))))).)).))))- ( -34.20, z-score = -1.94, R) >droPer1.super_80 159103 92 - 286535 ------AAAUUGCAGCAAUUGCAGGAGA--------GGUCUCACUUGGGGUGC-----ACC-U---CUCCCUCCGGGCUUAUGCAAAGCAAAUUGGUUUAUGCAUGAGGCGUCGG- ------....(((((...)))))(((((--------(((..(((.....))).-----)))-)---))))..(((((((((((((((((......)))).))))))).)).))))- ( -34.20, z-score = -1.94, R) >droWil1.scaffold_180698 4090211 104 - 11422946 ------AAAUUGCAGCAAUUGGAUAGGUCUAAACCAAGCUGAAACCGAAACCGUCGCCGUCUCGCACUGCCCUUUAGCUUAUGCAAAGCAAAUUGGUUUAUGCAUGAGGC------ ------.....((((...(((((....))))).....((.((...(((.....)))...))..)).)))).......((((((((((((......)))).))))))))..------ ( -22.30, z-score = 0.89, R) >droVir3.scaffold_13049 7946128 115 - 25233164 AACUUGCUGAAUUCGGAGUUAGACGCGUCAUCCCGGAAUUCGUUC-CCCAUACUUAUUUGACCCUAGUGCGCCUGAACUUAUGCAAAGCAAAUUGGUUUAUGCAUGAGGUGCCCAA ........(((((((((....((....)).)))..))))))....-....................(.((((((.....((((((((((......)))).)))))))))))).).. ( -23.60, z-score = 0.61, R) >droMoj3.scaffold_6680 2445609 99 - 24764193 ----------------AGUUAGCCGCGUCAUCCUGGCAUUCCUUC-UCCACACUUCUUUGACCCUGGUGCGCCUGAGCUUAGGCAACGCAAAUUGGCUUAUGCAUGAGCUGCCCAA ----------------.....((((.(....).))))........-..................(((.(((((((....)))))..........((((((....))))))))))). ( -22.10, z-score = 1.54, R) >droGri2.scaffold_15110 14896895 116 + 24565398 AAAGUGCAGCAAUCGGAGUUGGACGCGUCAUCCUGGAAUACGUUUGCUCCUACUUAUUUUACCCUAGUGCGCCUCAGCUUAUGCAAAACAAAUUGGUUUAUGCAUGAGGCUCCCAA ..............(((((.((.((((..(...(((((((.((........)).)))))))...)..))))))....((((((((((((......)))).)))))))))))))... ( -28.50, z-score = -0.08, R) >consensus ______AAAUUGCUGCAAUUGGAGAGGCUCUCUGGGGGUAUCCCCCAAAAUCCC__CUAUC_C___CUGCCCCCUG_CUUAUGCAAAGCAAAUUGGUUUAUGCAUGAGGCGGCGA_ .............................................................................((((((((((((......)))).))))))))........ (-11.14 = -11.16 + 0.01)
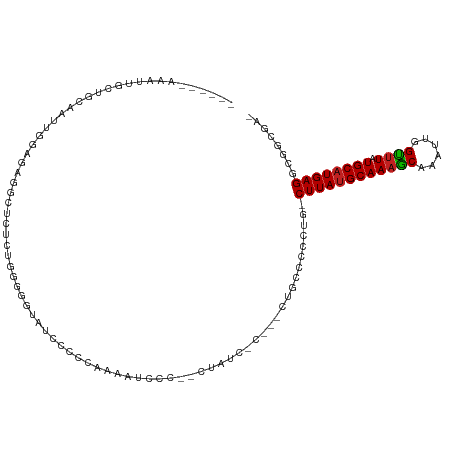
| Location | 22,472,784 – 22,472,888 |
|---|---|
| Length | 104 |
| Sequences | 12 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 60.58 |
| Shannon entropy | 0.84500 |
| G+C content | 0.51321 |
| Mean single sequence MFE | -32.38 |
| Consensus MFE | -11.24 |
| Energy contribution | -11.26 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.27 |
| Structure conservation index | 0.35 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.15 |
| SVM RNA-class probability | 0.900673 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 22472784 104 + 24543557 ------CAAUUGGAGAGUCUCUCUGGGGGAAUCCCCCAAAAUCC----CCCCU-CUCCCUGCCCGCUGCUUAUGCAAAGCAAAUUGGUUUAUGCAUGAGGCGGC-GAACAGACGAA ------.....((((((.......((((((...........)))----)))))-))))(((..(((((((((((((((((......)))).))))))).)))))-)..)))..... ( -41.61, z-score = -3.50, R) >droSim1.chr3L 21782566 104 + 22553184 ------CAAUUGGAGAGUCUCUCUGGGGGAAUCCCCCAAAAUCC----CCCCU-AUCCCUGCCUGCUGCUUAUGCAAAGCAAAUUGGUUUAUGCAUGAGGCGGC-UAACAGACGAA ------.....((((.....))))((((((...........)))----)))..-....(((...((((((((((((((((......)))).))))))).)))))-...)))..... ( -34.60, z-score = -1.75, R) >droSec1.super_11 2408587 104 + 2888827 ------CAAUUGGAGAGUCUCUCUGGGGGAAUCCCCAAAAAUCC----CACCU-AUCCCUGCCCCCUGCUUAUGCAAAGCAAAUUGGUUUAUGCAUGAGGCGGC-UAACAGACGAA ------....(((((((...))))((((....)))).......)----))...-....(((....(((((((((((((((......)))).))))))).)))).-...)))..... ( -27.70, z-score = -0.03, R) >droYak2.chr3L_random 99254 105 + 4797643 ------CAAUUGGAGAGUAUCUCUGGGGGAAUCCCCAAAAACCC----CCUAUGCCGCCUGCCCGCUGCUUAUGCAAAGCAAAUUGGUUUAUGCAUGAGGCGGC-GAACAGACGAA ------.....((((.....))))(((((............)))----)).....((.(((..(((((((((((((((((......)))).))))))).)))))-)..))).)).. ( -38.10, z-score = -2.47, R) >droEre2.scaffold_4784 22142242 101 + 25762168 ------CAAUUGGAGAGUCUCUCUGGGGGAAUGCCCCCAAACCC----CCUAU----CCCUGCCGCUGCUUAUGCAAAGCAAAUUGGUUUAUGCAUGAGCCGGC-GAGCAGACCAA ------...((((((((...))))(((((....)))))......----.....----..(((((((((((((((((((((......)))).)))))))).))))-).)))).)))) ( -42.40, z-score = -3.88, R) >droAna3.scaffold_13337 22210641 94 + 23293914 ------CAAUUGGAGG----CAGUGUGGUGUUGCC----AACUG----CCCAU---UCCUAACUCCUGCUUAUGCAAAGCAAAUUGGUUUAUGCAUGAGGCGGC-GGACAGACGAA ------.....(((((----((((.(((.....))----)))))----))...---)))...((.(((((((((((((((......)))).))))))).)))).-))......... ( -31.30, z-score = -1.17, R) >dp4.chrXR_group8 140319 92 + 9212921 ------CAAUUGCAG---------GAGAGGUCUCACUUGGGGUG----CACCU----CUCCCUCCGGGCUUAUGCAAAGCAAAUUGGUUUAUGCAUGAGGCGUC-GGACAGACGCA ------........(---------(((((((..(((.....)))----.))))----)))).......((((((((((((......)))).))))))))(((((-.....))))). ( -36.70, z-score = -2.57, R) >droPer1.super_80 159112 92 - 286535 ------CAAUUGCAG---------GAGAGGUCUCACUUGGGGUG----CACCU----CUCCCUCCGGGCUUAUGCAAAGCAAAUUGGUUUAUGCAUGAGGCGUC-GGACAGACGCA ------........(---------(((((((..(((.....)))----.))))----)))).......((((((((((((......)))).))))))))(((((-.....))))). ( -36.70, z-score = -2.57, R) >droWil1.scaffold_180698 4090220 95 - 11422946 ------CAAUUGGAUAGGUCUAAACCAAGCUGAAACCGAAACCGUCGCCGUCUCGCACUGCCCUUUAGCUUAUGCAAAGCAAAUUGGUUUAUGCAUGAGGC--------------- ------.....((...(((....)))..((.((...(((.....)))...))..)).....)).....((((((((((((......)))).))))))))..--------------- ( -18.90, z-score = 0.98, R) >droVir3.scaffold_13049 7946137 115 - 25233164 AAUUCGGAGUUAGACGCGUCAUCCCGGAAUUCGUUC-CCCAUACUUAUUUGACCCUAGUGCGCCUGAACUUAUGCAAAGCAAAUUGGUUUAUGCAUGAGGUGCCCAAACGGACAGA ..(((((.(...((....))..)))))).((((((.-..((........))......(.((((((.....((((((((((......)))).)))))))))))).).)))))).... ( -24.10, z-score = 0.79, R) >droMoj3.scaffold_6680 2445609 108 - 24764193 -------AGUUAGCCGCGUCAUCCUGGCAUUCCUUC-UCCACACUUCUUUGACCCUGGUGCGCCUGAGCUUAGGCAACGCAAAUUGGCUUAUGCAUGAGCUGCCCAAGAGGACGGA -------.....((((.(....).))))..((((((-((..((......))....(((.(((((((....)))))..........((((((....))))))))))).))))).))) ( -28.00, z-score = 1.37, R) >droGri2.scaffold_15110 14896904 116 + 24565398 CAAUCGGAGUUGGACGCGUCAUCCUGGAAUACGUUUGCUCCUACUUAUUUUACCCUAGUGCGCCUCAGCUUAUGCAAAACAAAUUGGUUUAUGCAUGAGGCUCCCAAACGGACAGA .....(((((.((.((((..(...(((((((.((........)).)))))))...)..))))))....((((((((((((......)))).)))))))))))))............ ( -28.50, z-score = -0.40, R) >consensus ______CAAUUGGAGAGUCUCUCUGGGGGAAUCCCCCAAAACCC____CCCAU_CU_CCUCCCCCCUGCUUAUGCAAAGCAAAUUGGUUUAUGCAUGAGGCGGC_GAACAGACGAA ....................................................................((((((((((((......)))).))))))))................. (-11.24 = -11.26 + 0.01)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:44:56 2011