| Sequence ID | dm3.chr3L |
|---|---|
| Location | 22,467,204 – 22,467,301 |
| Length | 97 |
| Max. P | 0.690420 |

| Location | 22,467,204 – 22,467,301 |
|---|---|
| Length | 97 |
| Sequences | 7 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 63.43 |
| Shannon entropy | 0.65845 |
| G+C content | 0.49474 |
| Mean single sequence MFE | -30.89 |
| Consensus MFE | -9.78 |
| Energy contribution | -10.30 |
| Covariance contribution | 0.51 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.32 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.690420 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
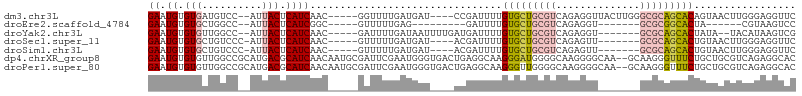
>dm3.chr3L 22467204 97 + 24543557 GAAUGUGUGAUGUCC--AUUACUCAUCAAC-----GGUUUUGAUGAU----CCGAUUUUGUGCUGCGUCAGAGGUUACUUGGGCGCAGCACAGUAACUUGGGAGGUUC ((((.((((((....--)))))(((((((.-----....)))))))(----((((..((((((((((((.(((....))).))))))))))))....)))))).)))) ( -36.80, z-score = -3.55, R) >droEre2.scaffold_4784 22136781 79 + 25762168 GAAUGUGUGCUGGCC--AUUACUCAUCGGC-----GUUUUUGAG---------GAUUUUGUGCUGCGUCAGAGGU-------GCGCGGCACUA------CGUAAGUCC ........(((((..--........)))))-----........(---------(((((.(((((((((.......-------)))))))))..------...)))))) ( -22.80, z-score = 0.25, R) >droYak2.chr3L 1253656 92 - 24197627 GAAUGUGUGUUGGCC--AUUACUCAUCAAC-----GAUUUUGAUAAUUUUGAUGAUUUUGUGCUGCGUCAGAGGU-------GCGCAGCACUAUA--UACAUAAGUCG ..((((((((.....--.....(((((((.-----((((.....)))))))))))....(((((((((.......-------))))))))).)))--)))))...... ( -27.90, z-score = -2.26, R) >droSec1.super_11 2403072 91 + 2888827 GAAUGUGUGCUGUCCC-AUUACUCAUCAAC-----GUUUUUGAUGAU----ACGAUUUUGUGCUGCGUCAGAGUU-------GCGCAGCACUGUAACUUGGGAGGUUC ........(((.((((-((((((((((((.-----....))))))).----........(((((((((.......-------))))))))).))))..)))))))).. ( -30.80, z-score = -2.53, R) >droSim1.chr3L 21776306 91 + 22553184 GAAUGUGUGCUGUCCC-AUUACUCAUCAAC-----GUUUUUGAUGAU----ACGAUUUUGUGCUGCGUCAGAGUU-------GCGCAGCACUGUAACUUGGGAGGUUC ........(((.((((-((((((((((((.-----....))))))).----........(((((((((.......-------))))))))).))))..)))))))).. ( -30.80, z-score = -2.53, R) >dp4.chrXR_group8 132393 106 + 9212921 GAAUGUGUGUUGGCCGCAUGACGCAUCAACAAUGCGAUUCGAAUGGGUGACUGAGGCAAGGGAUGGGGCAAGGGGCAA--GCAAGGGUUUCUGCUGCGUCAGAGGCAC ....((((.((.(((......(((((.....)))))........((....))..)))....((((.((((.(..((..--......))..))))).)))).)).)))) ( -33.20, z-score = -0.64, R) >droPer1.super_80 151199 106 - 286535 GAAUGUGUGUUGGCCGCAUGACGCAUCAACAAUGCGAUUCGAAUGGGUGACUGAGGCAAGGGUUGGGGCAAGGGGCAA--GCAAGGGUUUCUGCUGCGUCAGAGGCAC ............(((...((((((.(((((..(((..(((....((....))))))))...)))))((((.(..((..--......))..)))))))))))..))).. ( -33.90, z-score = -0.65, R) >consensus GAAUGUGUGCUGGCC__AUUACUCAUCAAC_____GAUUUUGAUGAU____ACGAUUUUGUGCUGCGUCAGAGGU_______GCGCAGCACUGUAACUUCGGAGGUUC ((.((.(((..........))).))))................................(((((((((..............)))))))))................. ( -9.78 = -10.30 + 0.51)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:44:53 2011