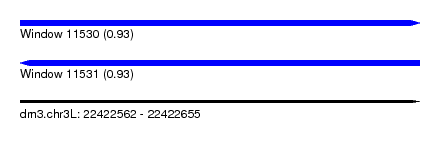
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 22,422,562 – 22,422,655 |
| Length | 93 |
| Max. P | 0.933351 |

| Location | 22,422,562 – 22,422,655 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 79.49 |
| Shannon entropy | 0.30400 |
| G+C content | 0.44907 |
| Mean single sequence MFE | -20.57 |
| Consensus MFE | -15.32 |
| Energy contribution | -15.88 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.74 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.38 |
| SVM RNA-class probability | 0.933351 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
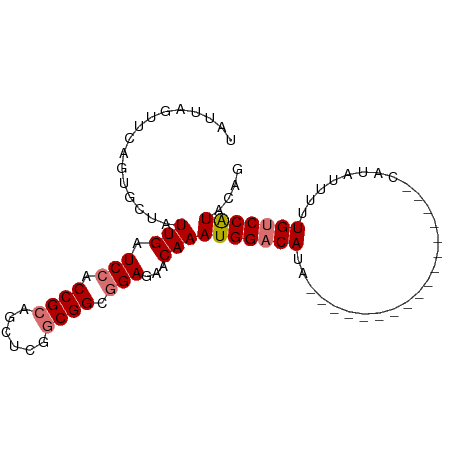
>dm3.chr3L 22422562 93 + 24543557 UAUUAGUGCAGUGCUAUUGAUCCACCGCAGCUCGGCGGCGGAGAACAAAUGGACAUAUUAUACAUACAUACGUACAUAUUUCUGUCCAUACAG ...((((.....))))(((.(((.((((......)))).)))...)))(((((((....(((..(((....)))..)))...))))))).... ( -23.60, z-score = -1.81, R) >droEre2.scaffold_4784 22093192 70 + 25762168 UACU-GUCCAGGUCUAUUGAUCCACCGCACUUCGUCGGCAGAGAACAAAUGGACAU----------------------UUUUUGACCGUACAG (((.-(((...(((((((..((..((((.....).)))..)).....)))))))..----------------------.....))).)))... ( -13.70, z-score = -0.25, R) >droSec1.super_11 2359358 76 + 2888827 UAUUAGUUCAGUGCUAUUGAUCCACCGCAGCUCGGCGGCGGAGAACAAAUGGACAUA-----------------CAUAUUUUUGUCCAUACAG .....((((....(....).(((.((((......)))).)))))))..(((((((..-----------------........))))))).... ( -22.50, z-score = -3.11, R) >droSim1.chr3L 21731413 76 + 22553184 UAUUAGUUCAGUGCUAUUGAUCCACCGCAGCUCGGCGGCGGAGAACAAAUGGACAUA-----------------CAUAUUUUUGUCCAUACAG .....((((....(....).(((.((((......)))).)))))))..(((((((..-----------------........))))))).... ( -22.50, z-score = -3.11, R) >consensus UAUUAGUUCAGUGCUAUUGAUCCACCGCAGCUCGGCGGCGGAGAACAAAUGGACAUA_________________CAUAUUUUUGUCCAUACAG ................(((.(((.((((......)))).)))...)))(((((((...........................))))))).... (-15.32 = -15.88 + 0.56)

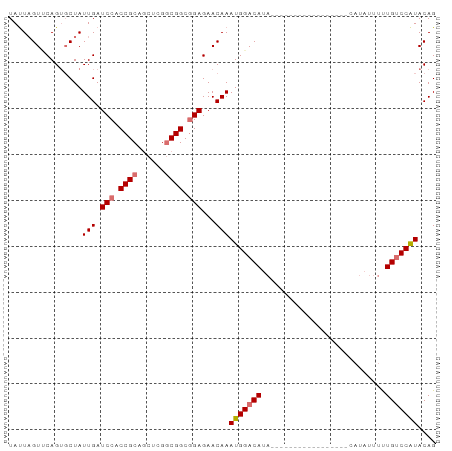
| Location | 22,422,562 – 22,422,655 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 79.49 |
| Shannon entropy | 0.30400 |
| G+C content | 0.44907 |
| Mean single sequence MFE | -23.15 |
| Consensus MFE | -16.92 |
| Energy contribution | -17.48 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.73 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.34 |
| SVM RNA-class probability | 0.928206 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
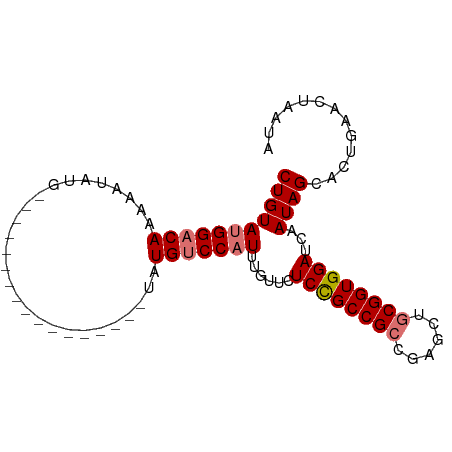
>dm3.chr3L 22422562 93 - 24543557 CUGUAUGGACAGAAAUAUGUACGUAUGUAUGUAUAAUAUGUCCAUUUGUUCUCCGCCGCCGAGCUGCGGUGGAUCAAUAGCACUGCACUAAUA .((((((((((...((((((((....))))))))....)))))).(((((.((((((((......))))))))..)))))...))))...... ( -29.50, z-score = -2.38, R) >droEre2.scaffold_4784 22093192 70 - 25762168 CUGUACGGUCAAAAA----------------------AUGUCCAUUUGUUCUCUGCCGACGAAGUGCGGUGGAUCAAUAGACCUGGAC-AGUA ...............----------------------.((((((((((((.((..(((........)))..))..))))))..)))))-)... ( -18.90, z-score = -1.18, R) >droSec1.super_11 2359358 76 - 2888827 CUGUAUGGACAAAAAUAUG-----------------UAUGUCCAUUUGUUCUCCGCCGCCGAGCUGCGGUGGAUCAAUAGCACUGAACUAAUA .((((((((((........-----------------..)))))))(((...((((((((......)))))))).)))..)))........... ( -22.10, z-score = -2.38, R) >droSim1.chr3L 21731413 76 - 22553184 CUGUAUGGACAAAAAUAUG-----------------UAUGUCCAUUUGUUCUCCGCCGCCGAGCUGCGGUGGAUCAAUAGCACUGAACUAAUA .((((((((((........-----------------..)))))))(((...((((((((......)))))))).)))..)))........... ( -22.10, z-score = -2.38, R) >consensus CUGUAUGGACAAAAAUAUG_________________UAUGUCCAUUUGUUCUCCGCCGCCGAGCUGCGGUGGAUCAAUAGCACUGAACUAAUA (((((((((((...........................)))))))......((((((((......))))))))...))))............. (-16.92 = -17.48 + 0.56)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:44:49 2011