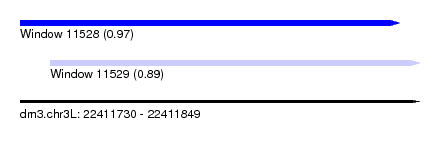
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 22,411,730 – 22,411,849 |
| Length | 119 |
| Max. P | 0.967834 |

| Location | 22,411,730 – 22,411,843 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 61.48 |
| Shannon entropy | 0.70018 |
| G+C content | 0.37490 |
| Mean single sequence MFE | -24.75 |
| Consensus MFE | -8.07 |
| Energy contribution | -9.51 |
| Covariance contribution | 1.44 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.33 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.79 |
| SVM RNA-class probability | 0.967834 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
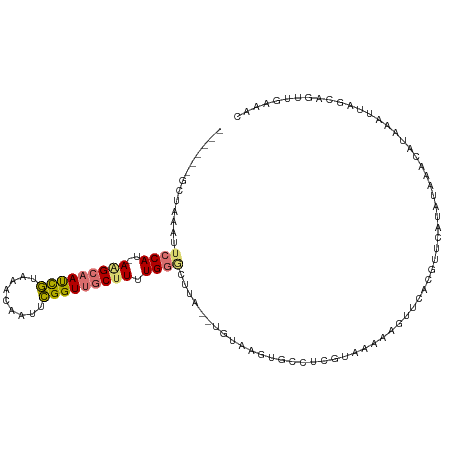
>dm3.chr3L 22411730 113 + 24543557 ----ACUAGAAAAGCAGAGGCUAAAUUCCAUAAGCAAUCGUAAACAAUUCGGUUGCUUUUGGGCUUAUGUAAGUGCCUCGUAAAAAGUUCUCCUUCAUGUAAACAUAAAUUAGCAGU- ----.........((.(((((......(((.(((((((((.........))))))))).)))((((....))))))))).................(((....)))......))...- ( -28.40, z-score = -2.82, R) >droSim1.chr3L 21719775 113 + 22553184 ----ACUAGAAACGCAGAGGCUAAAUUCCAUAGGCAAUCGUAAACAAUUCGGUUGCUUUUGGGCUUAUGUAAGUGCCUCGUAAAAAGUUCAUGUUCAUAUAAACAUAAAUUAGCAGU- ----.........((.(((((......(((.(((((((((.........))))))))).)))((((....)))))))))......((((.(((((......))))).)))).))...- ( -31.00, z-score = -3.52, R) >droSec1.super_11 2348297 99 + 2888827 ----ACUAGAAACGCAGA--------------GGCAAUCGUAAACAAUUCGGUUGCUUUUGGGCUUAUGUAAGUGCCUCGUAAAAAGUUCAUGUUCAUAUAAACAUGAAUUAGCAGU- ----.......(((((((--------------((((((((.........))))))))))))(((..........))).)))....((((((((((......))))))))))......- ( -28.40, z-score = -3.32, R) >droEre2.scaffold_4784 22082234 85 + 25762168 ----UCCAGCAAUGCAGAGGCUAAAUUCCACCAGAAAUCGUAAACCAUGCGGUUGCUUUUGGUUAUGAGUAAGUGCCUCGUAAACAGUU----------------------------- ----........(((.(((((...((((.(((((((((((((.....))))))...)))))))...))))....)))))))).......----------------------------- ( -21.90, z-score = -1.67, R) >droPer1.super_80 88892 103 - 286535 ACACACU-GAAAUGUAAA-----AAUCCAUCAAGAAACUAUUAAAAAUUUGGUGGCCUUUGG---------AGUGAGUCGUAAAUAAUCUAGCCGUUGCCUCUAAUGAAGUGAAAAUU ...((((-..........-----...((((((((.............))))))))...((((---------((.(...((.............))...)))))))...))))...... ( -14.04, z-score = 0.52, R) >consensus ____ACUAGAAACGCAGAGGCUAAAUUCCACAAGCAAUCGUAAACAAUUCGGUUGCUUUUGGGCUUAUGUAAGUGCCUCGUAAAAAGUUCAUCUUCAUAUAAACAUAAAUUAGCAGU_ .............(((...........(((.(((((((((.........))))))))).)))...........))).......................................... ( -8.07 = -9.51 + 1.44)


| Location | 22,411,739 – 22,411,849 |
|---|---|
| Length | 110 |
| Sequences | 4 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 67.27 |
| Shannon entropy | 0.51935 |
| G+C content | 0.36522 |
| Mean single sequence MFE | -24.66 |
| Consensus MFE | -9.25 |
| Energy contribution | -9.50 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.42 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.09 |
| SVM RNA-class probability | 0.889096 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 22411739 110 + 24543557 GCAGAGGCUAAAUUCCAU-AAGCAAUCGUAAACAAUUCGGUUGCUUUUGGGCUUA--UGUAAGUGCCUCGUAAAAAGUUCUCCUUCAUGUAAACAUAAAUUAGCAGUUGAAAC ((.(((((......(((.-(((((((((.........))))))))).)))((((.--...))))))))).................(((....)))......))......... ( -28.20, z-score = -2.94, R) >droSim1.chr3L 21719784 110 + 22553184 GCAGAGGCUAAAUUCCAU-AGGCAAUCGUAAACAAUUCGGUUGCUUUUGGGCUUA--UGUAAGUGCCUCGUAAAAAGUUCAUGUUCAUAUAAACAUAAAUUAGCAGUUGAAAC ((.(((((......(((.-(((((((((.........))))))))).)))((((.--...)))))))))......((((.(((((......))))).)))).))......... ( -30.70, z-score = -3.77, R) >droSec1.super_11 2348306 96 + 2888827 --------------GCAG-AGGCAAUCGUAAACAAUUCGGUUGCUUUUGGGCUUA--UGUAAGUGCCUCGUAAAAAGUUCAUGUUCAUAUAAACAUGAAUUAGCAGUUGAAAC --------------.(((-(((((((((.........))))))))))))(((...--.......)))........((((((((((......))))))))))............ ( -27.40, z-score = -3.26, R) >droPer1.super_80 88904 103 - 286535 ------GUAAAAAUCCAUCAAGAAACUAUUAAAAAUUUGGUGGCCUUUGGAGUGAGUCGUAAAUAAUCU---AGCCGUUGCCUCUAAUG-AAGUGAAAAUUGACCCAGAAAAU ------........((((((((.............))))))))..(((((.((.(((............---.....(..(.((....)-).)..)..))).))))))).... ( -12.33, z-score = 0.82, R) >consensus ______GCUAAAUUCCAU_AAGCAAUCGUAAACAAUUCGGUUGCUUUUGGGCUUA__UGUAAGUGCCUCGUAAAAAGUUCACGUUCAUAUAAACAUAAAUUAGCAGUUGAAAC .............((((...((((((((.........))))))))..)))).............................................................. ( -9.25 = -9.50 + 0.25)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:44:48 2011