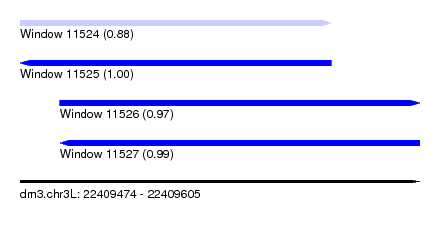
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 22,409,474 – 22,409,605 |
| Length | 131 |
| Max. P | 0.998137 |

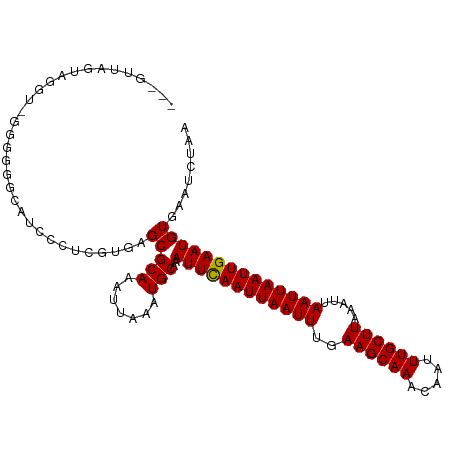
| Location | 22,409,474 – 22,409,576 |
|---|---|
| Length | 102 |
| Sequences | 9 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 87.61 |
| Shannon entropy | 0.26805 |
| G+C content | 0.33375 |
| Mean single sequence MFE | -26.42 |
| Consensus MFE | -16.76 |
| Energy contribution | -16.67 |
| Covariance contribution | -0.10 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.04 |
| SVM RNA-class probability | 0.879614 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr3L 22409474 102 + 24543557 ---GUUAGUGGG--GUAGGGCAUCCCUCGUGAGCGCAAAUUAAAUGCAAAUUCAAUUAAUUUGAAGCAAACAAUUUGCUUAAAUUAAUUAAUUGAAUGUGAAUCUAA ---(((.(.(((--((.....))))).)...)))(((.......)))..((((((((((((..((((((.....)))))).....)))))))))))).......... ( -24.90, z-score = -2.27, R) >droSim1.chr3L 21717536 104 + 22553184 ---GCUAGUGGGUUGUGGGGCAUCCCUCGUAAGCGCAAAUUAAAUGCAAAUUUAAUUAAUUUGAAGCAAACAAUUUGCUUAAAUUAAUUAAUUGAAUGUGAAUCUAA ---(((.(.(((.((.....)).))).)...)))(((.......)))..((((((((((((..((((((.....)))))).....)))))))))))).......... ( -24.10, z-score = -1.69, R) >droSec1.super_11 2346047 104 + 2888827 ---GUUAGUGGGUUGUGGGGCAUCCCUCGUGAGCGCAAAUUAAAUGCAAAUUCAAUUAAUUUGAAGCAAACAAUUUGCUUAAAUUAAUUAAUUGAAUGUGAAUCUAA ---.......(.(..(((((....)))))..).)(((.......)))..((((((((((((..((((((.....)))))).....)))))))))))).......... ( -25.40, z-score = -2.08, R) >droYak2.chr3L 22838473 102 + 24197627 ---GUUAGUAGG--GGGCGGUAUCCCUCGUGAGCGCAAAUUAAAUGCAAAUUCAAUUAAUUUGAAGCAAACAAUUUGCUUAAAUUAAUUAAUUGAAUGUGAAUCUAA ---(((....((--(((......)))))...)))(((.......)))..((((((((((((..((((((.....)))))).....)))))))))))).......... ( -25.70, z-score = -2.68, R) >droEre2.scaffold_4784 22079999 102 + 25762168 ---GUUAGUAGG--GGGCGGCGUCCCUCGUGAGCGCAAAUUAAAUGCAAAUUCAAUUAAUUUGAAGCAAACAAUUUGCUUAAAUUAAUUAAUUGAAUGUGAAUCUAA ---(((.(..((--((((...))))))..).)))(((.......)))..((((((((((((..((((((.....)))))).....)))))))))))).......... ( -27.00, z-score = -2.64, R) >droAna3.scaffold_13337 22148080 93 + 23293914 --------------GGGCGGCGGCCCUUGCGGGCGCAAAUUAAAUGCAAAUUCAAUUAAUUUGAAGCAAACAAUUUGCUUAAAUUAAUUAAUUGAAUGUGAAUCUAA --------------((((....))))((((....))))........((.((((((((((((..((((((.....)))))).....)))))))))))).))....... ( -26.50, z-score = -2.46, R) >dp4.chrXR_group8 68526 106 + 9212921 UGCCGCCGCCAGUCG-GGGGCGUCUCUCGUGAGCGCAAAUUAAAUGCAAAUUCAAUUAAUUUGAAGCAAACAAUUUGCUUAAAUUAAUUAAUUGAAUGUGAAUCUAA (((((((.((....)-).)))).(((....))).))).........((.((((((((((((..((((((.....)))))).....)))))))))))).))....... ( -30.70, z-score = -2.88, R) >droPer1.super_80 86099 106 - 286535 UGCCUCCGCCAGUCG-GGGGCGUCUCUCGUGAGCGCAAAUUAAAUGCAAAUUCAAUUAAUUUGAAGCAAACAAUUUGCUUAAAUUAAUUAAUUGAAUGUGAAUCUAA .(((((((.....))-)))))(.(((....))))(((.......)))..((((((((((((..((((((.....)))))).....)))))))))))).......... ( -31.90, z-score = -3.86, R) >droWil1.scaffold_180698 3996662 104 - 11422946 ---GUGAAUAUGUGUGUGUGAUUCUGUAGUGAGCGCAAAUUAAAUGCAAAUUCAAUUAAUUUGAAGCAAACAAUUUGCUUAAAUUAAUUAAUUGAAUGUGAAUCUAA ---.......(((((.((((.(((......)))))))......))))).((((((((((((..((((((.....)))))).....)))))))))))).......... ( -21.60, z-score = -1.44, R) >consensus ___GUUAGUAGGU_GGGGGGCAUCCCUCGUGAGCGCAAAUUAAAUGCAAAUUCAAUUAAUUUGAAGCAAACAAUUUGCUUAAAUUAAUUAAUUGAAUGUGAAUCUAA ................................(((((.......)))..((((((((((((..((((((.....)))))).....))))))))))))))........ (-16.76 = -16.67 + -0.10)

| Location | 22,409,474 – 22,409,576 |
|---|---|
| Length | 102 |
| Sequences | 9 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 87.61 |
| Shannon entropy | 0.26805 |
| G+C content | 0.33375 |
| Mean single sequence MFE | -24.44 |
| Consensus MFE | -20.54 |
| Energy contribution | -20.49 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.12 |
| Mean z-score | -3.39 |
| Structure conservation index | 0.84 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.27 |
| SVM RNA-class probability | 0.998137 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr3L 22409474 102 - 24543557 UUAGAUUCACAUUCAAUUAAUUAAUUUAAGCAAAUUGUUUGCUUCAAAUUAAUUGAAUUUGCAUUUAAUUUGCGCUCACGAGGGAUGCCCUAC--CCCACUAAC--- ((((((.((.((((((((((((.....((((((.....))))))..)))))))))))).)).))))))...((((((....))).))).....--.........--- ( -21.00, z-score = -2.72, R) >droSim1.chr3L 21717536 104 - 22553184 UUAGAUUCACAUUCAAUUAAUUAAUUUAAGCAAAUUGUUUGCUUCAAAUUAAUUAAAUUUGCAUUUAAUUUGCGCUUACGAGGGAUGCCCCACAACCCACUAGC--- ((((........((..(((((((((((((((((.....)))))).)))))))))))....(((.......)))......))(((.((.....)).))).)))).--- ( -21.10, z-score = -2.56, R) >droSec1.super_11 2346047 104 - 2888827 UUAGAUUCACAUUCAAUUAAUUAAUUUAAGCAAAUUGUUUGCUUCAAAUUAAUUGAAUUUGCAUUUAAUUUGCGCUCACGAGGGAUGCCCCACAACCCACUAAC--- ((((((.((.((((((((((((.....((((((.....))))))..)))))))))))).)).)))))).............(((.((.....)).)))......--- ( -22.20, z-score = -3.13, R) >droYak2.chr3L 22838473 102 - 24197627 UUAGAUUCACAUUCAAUUAAUUAAUUUAAGCAAAUUGUUUGCUUCAAAUUAAUUGAAUUUGCAUUUAAUUUGCGCUCACGAGGGAUACCGCCC--CCUACUAAC--- ((((((.((.((((((((((((.....((((((.....))))))..)))))))))))).)).))))))............((((........)--)))......--- ( -22.40, z-score = -3.41, R) >droEre2.scaffold_4784 22079999 102 - 25762168 UUAGAUUCACAUUCAAUUAAUUAAUUUAAGCAAAUUGUUUGCUUCAAAUUAAUUGAAUUUGCAUUUAAUUUGCGCUCACGAGGGACGCCGCCC--CCUACUAAC--- ((((((.((.((((((((((((.....((((((.....))))))..)))))))))))).)).))))))...((((((....))).))).....--.........--- ( -22.90, z-score = -3.11, R) >droAna3.scaffold_13337 22148080 93 - 23293914 UUAGAUUCACAUUCAAUUAAUUAAUUUAAGCAAAUUGUUUGCUUCAAAUUAAUUGAAUUUGCAUUUAAUUUGCGCCCGCAAGGGCCGCCGCCC-------------- ((((((.((.((((((((((((.....((((((.....))))))..)))))))))))).)).))))))...((((((....))).))).....-------------- ( -29.60, z-score = -4.65, R) >dp4.chrXR_group8 68526 106 - 9212921 UUAGAUUCACAUUCAAUUAAUUAAUUUAAGCAAAUUGUUUGCUUCAAAUUAAUUGAAUUUGCAUUUAAUUUGCGCUCACGAGAGACGCCCC-CGACUGGCGGCGGCA ((((((.((.((((((((((((.....((((((.....))))))..)))))))))))).)).))))))..(((.(((....))).((((.(-(....)).))))))) ( -30.40, z-score = -3.34, R) >droPer1.super_80 86099 106 + 286535 UUAGAUUCACAUUCAAUUAAUUAAUUUAAGCAAAUUGUUUGCUUCAAAUUAAUUGAAUUUGCAUUUAAUUUGCGCUCACGAGAGACGCCCC-CGACUGGCGGAGGCA ((((((.((.((((((((((((.....((((((.....))))))..)))))))))))).)).))))))..(((.(((.(....).((((..-.....)))))))))) ( -29.50, z-score = -3.37, R) >droWil1.scaffold_180698 3996662 104 + 11422946 UUAGAUUCACAUUCAAUUAAUUAAUUUAAGCAAAUUGUUUGCUUCAAAUUAAUUGAAUUUGCAUUUAAUUUGCGCUCACUACAGAAUCACACACACAUAUUCAC--- ...(((((........(((((((((((((((((.....)))))).)))))))))))....(((.......)))..........)))))................--- ( -20.90, z-score = -4.21, R) >consensus UUAGAUUCACAUUCAAUUAAUUAAUUUAAGCAAAUUGUUUGCUUCAAAUUAAUUGAAUUUGCAUUUAAUUUGCGCUCACGAGGGACGCCCCCC_ACCCACUAAC___ ((((((.((.((((((((((((.....((((((.....))))))..)))))))))))).)).))))))...((((((....))).)))................... (-20.54 = -20.49 + -0.05)

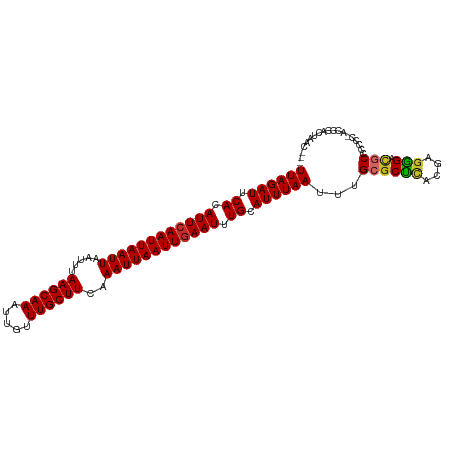
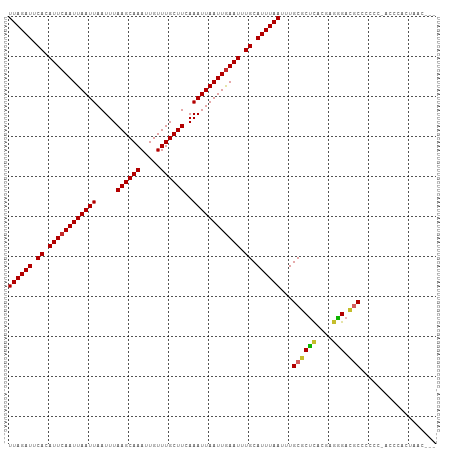
| Location | 22,409,487 – 22,409,605 |
|---|---|
| Length | 118 |
| Sequences | 12 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 92.20 |
| Shannon entropy | 0.18558 |
| G+C content | 0.32050 |
| Mean single sequence MFE | -27.91 |
| Consensus MFE | -22.37 |
| Energy contribution | -22.68 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.59 |
| Structure conservation index | 0.80 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.78 |
| SVM RNA-class probability | 0.967281 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr3L 22409487 118 + 24543557 GGCAUCCCU-CGUGAGCGCAAAUUAAAUGCAAAUUCAAUUAAUUUGAAGCAAACAAUUUGCUUAAAUUAAUUAAUUGAAUGUGAAUCUAAAGUGCUUCCACUUUAACGCUGAAGAGCUC .......((-(.(.((((...........((.((((((((((((..((((((.....)))))).....)))))))))))).))....(((((((....))))))).)))).).)))... ( -29.80, z-score = -3.17, R) >droSim1.chr3L 21717551 118 + 22553184 GGCAUCCCU-CGUAAGCGCAAAUUAAAUGCAAAUUUAAUUAAUUUGAAGCAAACAAUUUGCUUAAAUUAAUUAAUUGAAUGUGAAUCUAAAGUGCUUCCAAUUUAACGCUGAAGAGCUC .......((-(...((((.(((((....(((.((((((((((((..((((((.....)))))).....))))))))))))............)))....)))))..))))...)))... ( -23.72, z-score = -1.51, R) >droSec1.super_11 2346062 118 + 2888827 GGCAUCCCU-CGUGAGCGCAAAUUAAAUGCAAAUUCAAUUAAUUUGAAGCAAACAAUUUGCUUAAAUUAAUUAAUUGAAUGUGAAUCUAAAGUGCUUCCAAUUUAACGCUGAAGAGCUC .........-.(..(((((...(((.((.((.((((((((((((..((((((.....)))))).....)))))))))))).)).)).))).)))))..)........(((....))).. ( -26.30, z-score = -2.00, R) >droYak2.chr3L 22838486 118 + 24197627 GGUAUCCCU-CGUGAGCGCAAAUUAAAUGCAAAUUCAAUUAAUUUGAAGCAAACAAUUUGCUUAAAUUAAUUAAUUGAAUGUGAAUCUAAAGUGCUUCCAAUUUAACGCUGAAGAGCUC .........-.(..(((((...(((.((.((.((((((((((((..((((((.....)))))).....)))))))))))).)).)).))).)))))..)........(((....))).. ( -26.30, z-score = -2.41, R) >droEre2.scaffold_4784 22080012 118 + 25762168 GGCGUCCCU-CGUGAGCGCAAAUUAAAUGCAAAUUCAAUUAAUUUGAAGCAAACAAUUUGCUUAAAUUAAUUAAUUGAAUGUGAAUCUAAAGUGCUUCCAAUUUAACGCUGAAGAGCUC (((((....-.(..(((((...(((.((.((.((((((((((((..((((((.....)))))).....)))))))))))).)).)).))).)))))..)......)))))......... ( -28.70, z-score = -2.70, R) >droAna3.scaffold_13337 22148084 117 + 23293914 GGCGGCCCU-UGCGGGCGCAAAUUAAAUGCAAAUUCAAUUAAUUUGAAGCAAACAAUUUGCUUAAAUUAAUUAAUUGAAUGUGAAUCUAAAGUGCUUCCAAUUUAACGCUGAGACCCC- ((((((((.-...))))(((..(((.((.((.((((((((((((..((((((.....)))))).....)))))))))))).)).)).)))..)))...........))))........- ( -33.60, z-score = -3.59, R) >dp4.chrXR_group8 68543 118 + 9212921 GGCGUCUCU-CGUGAGCGCAAAUUAAAUGCAAAUUCAAUUAAUUUGAAGCAAACAAUUUGCUUAAAUUAAUUAAUUGAAUGUGAAUCUAAAGUGCUUCCAAUUUAACGCUGAAGAGCUC (((((....-.(..(((((...(((.((.((.((((((((((((..((((((.....)))))).....)))))))))))).)).)).))).)))))..)......)))))......... ( -28.70, z-score = -2.48, R) >droPer1.super_80 86116 118 - 286535 GGCGUCUCU-CGUGAGCGCAAAUUAAAUGCAAAUUCAAUUAAUUUGAAGCAAACAAUUUGCUUAAAUUAAUUAAUUGAAUGUGAAUCUAAAGUGCUUCCAAUUUAACGCUGAAGAGCUC (((((....-.(..(((((...(((.((.((.((((((((((((..((((((.....)))))).....)))))))))))).)).)).))).)))))..)......)))))......... ( -28.70, z-score = -2.48, R) >droWil1.scaffold_180698 3996677 118 - 11422946 UGAUUCUGU-AGUGAGCGCAAAUUAAAUGCAAAUUCAAUUAAUUUGAAGCAAACAAUUUGCUUAAAUUAAUUAAUUGAAUGUGAAUCUAAAGUGCUUCCAAUUUAACGCUGAAGAGGCG .........-.(..(((((...(((.((.((.((((((((((((..((((((.....)))))).....)))))))))))).)).)).))).)))))..).......((((.....)))) ( -27.70, z-score = -2.28, R) >droVir3.scaffold_13049 7876072 104 - 25233164 --------------AGCGCAAAUUAAAUGCAAAUUCAAUUAAUUUGAAGCAAACAAUUUGCUUAAAUUAAUUAAUUGAAUGUGAAUCUAAAGUGCUUCCAAUUUAACGCUGAAGAGCA- --------------(((((...(((.((.((.((((((((((((..((((((.....)))))).....)))))))))))).)).)).))).)))))...........(((....))).- ( -23.50, z-score = -1.57, R) >droMoj3.scaffold_6680 2362635 116 - 24764193 --UGUGUCU-CGUAAGCACAAAUUAAAUGCAAAUUCAAUUAAUUUGAAGCAAACAAUUUGCUUAAAUUAAUUAAUUAAAUGCGAAUCUAAAGUGUUUCCAAUUUAACGCUAAAGAGCUC --(((((..-.....))))).......((((((((.(((((((((((.((((.....)))))))))))))))))))...))))..(((..((((((........))))))..))).... ( -23.50, z-score = -2.26, R) >droGri2.scaffold_15110 14820160 118 + 24565398 UGCGUCUCUUCGUGAGCACAAAUUAAAUGCAAAUUCAAUUAAUUUGAAGCAAACAAUUUGCUUAAAUUAAUUAAUUGAAUGUGAAUCUAAAGUGCUUCCAAUUGAACGAUGAAGAGGC- ...(((((((((..(((((...(((.((.((.((((((((((((..((((((.....)))))).....)))))))))))).)).)).))).)))))..).(((....)))))))))))- ( -34.40, z-score = -4.60, R) >consensus GGCGUCCCU_CGUGAGCGCAAAUUAAAUGCAAAUUCAAUUAAUUUGAAGCAAACAAUUUGCUUAAAUUAAUUAAUUGAAUGUGAAUCUAAAGUGCUUCCAAUUUAACGCUGAAGAGCUC ...........(..(((((...(((.((.((.((((((((((((..((((((.....)))))).....)))))))))))).)).)).))).)))))..)........(((....))).. (-22.37 = -22.68 + 0.31)


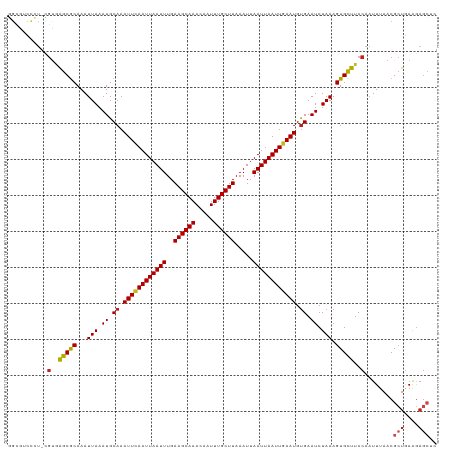
| Location | 22,409,487 – 22,409,605 |
|---|---|
| Length | 118 |
| Sequences | 12 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 92.20 |
| Shannon entropy | 0.18558 |
| G+C content | 0.32050 |
| Mean single sequence MFE | -31.02 |
| Consensus MFE | -23.61 |
| Energy contribution | -23.74 |
| Covariance contribution | 0.13 |
| Combinations/Pair | 1.09 |
| Mean z-score | -3.38 |
| Structure conservation index | 0.76 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.63 |
| SVM RNA-class probability | 0.993587 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr3L 22409487 118 - 24543557 GAGCUCUUCAGCGUUAAAGUGGAAGCACUUUAGAUUCACAUUCAAUUAAUUAAUUUAAGCAAAUUGUUUGCUUCAAAUUAAUUGAAUUUGCAUUUAAUUUGCGCUCACG-AGGGAUGCC ..........(((((...((((..(((..((((((.((.((((((((((((.....((((((.....))))))..)))))))))))).)).))))))..)))..)))).-...))))). ( -33.40, z-score = -3.43, R) >droSim1.chr3L 21717551 118 - 22553184 GAGCUCUUCAGCGUUAAAUUGGAAGCACUUUAGAUUCACAUUCAAUUAAUUAAUUUAAGCAAAUUGUUUGCUUCAAAUUAAUUAAAUUUGCAUUUAAUUUGCGCUUACG-AGGGAUGCC ...(((((.(((((.((((((((.(((.....((.......))..(((((((((((((((((.....)))))).)))))))))))...))).)))))))))))))...)-))))..... ( -30.30, z-score = -3.12, R) >droSec1.super_11 2346062 118 - 2888827 GAGCUCUUCAGCGUUAAAUUGGAAGCACUUUAGAUUCACAUUCAAUUAAUUAAUUUAAGCAAAUUGUUUGCUUCAAAUUAAUUGAAUUUGCAUUUAAUUUGCGCUCACG-AGGGAUGCC ..........(((((...(((((.(((..((((((.((.((((((((((((.....((((((.....))))))..)))))))))))).)).))))))..)))..)).))-)..))))). ( -31.40, z-score = -3.04, R) >droYak2.chr3L 22838486 118 - 24197627 GAGCUCUUCAGCGUUAAAUUGGAAGCACUUUAGAUUCACAUUCAAUUAAUUAAUUUAAGCAAAUUGUUUGCUUCAAAUUAAUUGAAUUUGCAUUUAAUUUGCGCUCACG-AGGGAUACC ...(((((.(((((.((((((((.(((.....((.......))..(((((((((((((((((.....)))))).)))))))))))...))).)))))))))))))...)-))))..... ( -30.40, z-score = -3.43, R) >droEre2.scaffold_4784 22080012 118 - 25762168 GAGCUCUUCAGCGUUAAAUUGGAAGCACUUUAGAUUCACAUUCAAUUAAUUAAUUUAAGCAAAUUGUUUGCUUCAAAUUAAUUGAAUUUGCAUUUAAUUUGCGCUCACG-AGGGACGCC ..........(((((...(((((.(((..((((((.((.((((((((((((.....((((((.....))))))..)))))))))))).)).))))))..)))..)).))-)..))))). ( -33.20, z-score = -3.86, R) >droAna3.scaffold_13337 22148084 117 - 23293914 -GGGGUCUCAGCGUUAAAUUGGAAGCACUUUAGAUUCACAUUCAAUUAAUUAAUUUAAGCAAAUUGUUUGCUUCAAAUUAAUUGAAUUUGCAUUUAAUUUGCGCCCGCA-AGGGCCGCC -.........(((...........(((..((((((.((.((((((((((((.....((((((.....))))))..)))))))))))).)).))))))..)))((((...-.))))))). ( -36.90, z-score = -4.06, R) >dp4.chrXR_group8 68543 118 - 9212921 GAGCUCUUCAGCGUUAAAUUGGAAGCACUUUAGAUUCACAUUCAAUUAAUUAAUUUAAGCAAAUUGUUUGCUUCAAAUUAAUUGAAUUUGCAUUUAAUUUGCGCUCACG-AGAGACGCC ..........(((((...(((((.(((..((((((.((.((((((((((((.....((((((.....))))))..)))))))))))).)).))))))..)))..)).))-)..))))). ( -32.10, z-score = -3.55, R) >droPer1.super_80 86116 118 + 286535 GAGCUCUUCAGCGUUAAAUUGGAAGCACUUUAGAUUCACAUUCAAUUAAUUAAUUUAAGCAAAUUGUUUGCUUCAAAUUAAUUGAAUUUGCAUUUAAUUUGCGCUCACG-AGAGACGCC ..........(((((...(((((.(((..((((((.((.((((((((((((.....((((((.....))))))..)))))))))))).)).))))))..)))..)).))-)..))))). ( -32.10, z-score = -3.55, R) >droWil1.scaffold_180698 3996677 118 + 11422946 CGCCUCUUCAGCGUUAAAUUGGAAGCACUUUAGAUUCACAUUCAAUUAAUUAAUUUAAGCAAAUUGUUUGCUUCAAAUUAAUUGAAUUUGCAUUUAAUUUGCGCUCACU-ACAGAAUCA .........(((((.((((((((.(((.....((.......))..(((((((((((((((((.....)))))).)))))))))))...))).)))))))))))))....-......... ( -26.10, z-score = -2.81, R) >droVir3.scaffold_13049 7876072 104 + 25233164 -UGCUCUUCAGCGUUAAAUUGGAAGCACUUUAGAUUCACAUUCAAUUAAUUAAUUUAAGCAAAUUGUUUGCUUCAAAUUAAUUGAAUUUGCAUUUAAUUUGCGCU-------------- -........(((((.((((((((.(((.....((.......))..(((((((((((((((((.....)))))).)))))))))))...))).)))))))))))))-------------- ( -26.00, z-score = -2.40, R) >droMoj3.scaffold_6680 2362635 116 + 24764193 GAGCUCUUUAGCGUUAAAUUGGAAACACUUUAGAUUCGCAUUUAAUUAAUUAAUUUAAGCAAAUUGUUUGCUUCAAAUUAAUUGAAUUUGCAUUUAAUUUGUGCUUACG-AGACACA-- ..((((...(((((((((.((....)).)))))....(((.....(((((((((((((((((.....)))))).)))))))))))...)))..........))))...)-)).)...-- ( -26.40, z-score = -2.46, R) >droGri2.scaffold_15110 14820160 118 - 24565398 -GCCUCUUCAUCGUUCAAUUGGAAGCACUUUAGAUUCACAUUCAAUUAAUUAAUUUAAGCAAAUUGUUUGCUUCAAAUUAAUUGAAUUUGCAUUUAAUUUGUGCUCACGAAGAGACGCA -((((((((....(((....)))(((((.((((((.((.((((((((((((.....((((((.....))))))..)))))))))))).)).))))))...)))))...))))))..)). ( -33.90, z-score = -4.81, R) >consensus GAGCUCUUCAGCGUUAAAUUGGAAGCACUUUAGAUUCACAUUCAAUUAAUUAAUUUAAGCAAAUUGUUUGCUUCAAAUUAAUUGAAUUUGCAUUUAAUUUGCGCUCACG_AGGGACGCC .........(((((.((((((((.(((.....((.......))..(((((((((((((((((.....)))))).)))))))))))...))).))))))))))))).............. (-23.61 = -23.74 + 0.13)

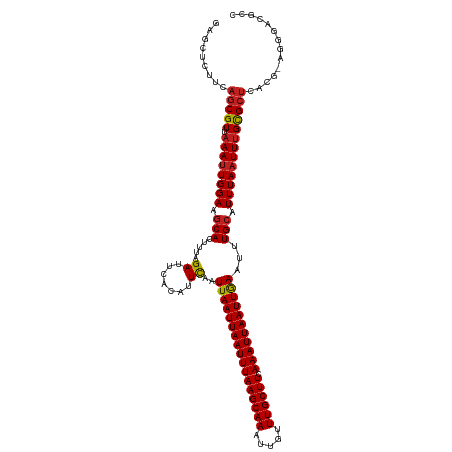
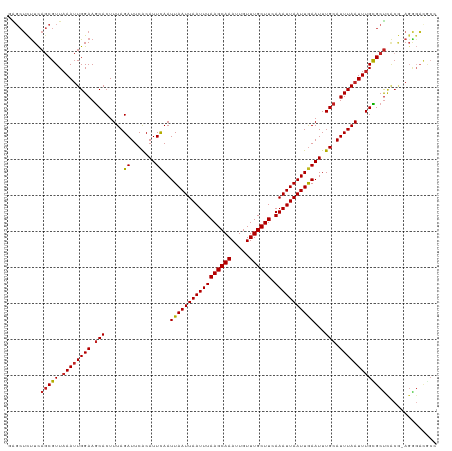
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:44:46 2011