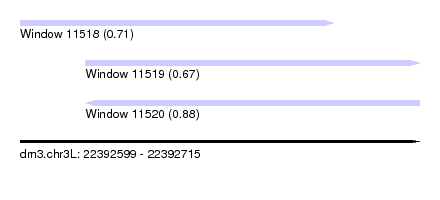
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 22,392,599 – 22,392,715 |
| Length | 116 |
| Max. P | 0.875359 |

| Location | 22,392,599 – 22,392,690 |
|---|---|
| Length | 91 |
| Sequences | 3 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 77.36 |
| Shannon entropy | 0.30329 |
| G+C content | 0.49484 |
| Mean single sequence MFE | -26.63 |
| Consensus MFE | -23.03 |
| Energy contribution | -23.03 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.13 |
| Structure conservation index | 0.86 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.713477 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
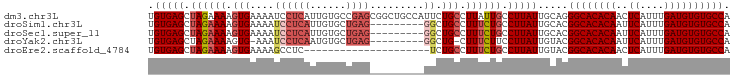
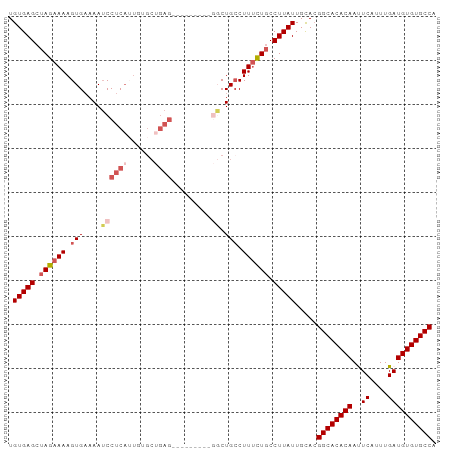
>dm3.chr3L 22392599 91 + 24543557 ------------------UAACUCAUAUACCCAGCCUUGGCACACAUCAAAUGAGUUGUGUGCCCUGCAAUAAGGCAAUAAGGCAGAAUGGCAGCCGCUCGGCACAAUG ------------------............((((((((((((((((.(......).)))))))).(((......)))..)))))....)))..(((....)))...... ( -27.70, z-score = -1.28, R) >droSim1.chr3L 21703779 100 + 22553184 UGACUCCUAUUCCCAGCCUGAUUCAUAUACCCAGCCUUGGCACACAUCAAAUGAAUUGUGUGCCGUGCAAUAAGGCAGAAAGGCAG---------CCCUCAGCACAAUG .................................(((((((((((((((....))..)))))))).(((......)))..))))).(---------(.....))...... ( -24.10, z-score = -0.73, R) >droSec1.super_11 2329130 100 + 2888827 UGACUCCUAUUCCCAGCCUGGUUCAUAUACCCAGCCUUGGCACACAUCAAAUGAAUUGUGUGCCGUGCAAUAAGGCAGAAAGGCAG---------CCCUCAGCACAAUG ((((((((.(((...(((((((......)))..((..(((((((((((....))..))))))))).))....)))).)))))).))---------...)))........ ( -28.10, z-score = -1.38, R) >consensus UGACUCCUAUUCCCAGCCUGAUUCAUAUACCCAGCCUUGGCACACAUCAAAUGAAUUGUGUGCCGUGCAAUAAGGCAGAAAGGCAG_________CCCUCAGCACAAUG .................................(((((((((((((((....))..)))))))).(((......)))..)))))......................... (-23.03 = -23.03 + 0.00)


| Location | 22,392,618 – 22,392,715 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 84.83 |
| Shannon entropy | 0.25849 |
| G+C content | 0.45218 |
| Mean single sequence MFE | -26.32 |
| Consensus MFE | -17.41 |
| Energy contribution | -18.74 |
| Covariance contribution | 1.33 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.669871 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 22392618 97 + 24543557 UGGCACACAUCAAAUGAGUUGUGUGCCCUGCAAUAAGGCAAUAAGGCAGAAUGGCAGCCGCUCGGCACAAUGAGGAUUUUCACUUUUCUAGCUCACA .((((((((.(......).)))))))).(((......)))....(((((((.((..(((....)))....((((....)))))).)))).))).... ( -25.80, z-score = -0.02, R) >droSim1.chr3L 21703816 88 + 22553184 UGGCACACAUCAAAUGAAUUGUGUGCCGUGCAAUAAGGCAGAAAGGCAGCC---------CUCAGCACAAUGAGGAUUUUCACUUUUCUAGCUCACA .((((((((((....))..))))))))(((......((((((((((....(---------((((......))))).......))))))).)))))). ( -29.60, z-score = -2.91, R) >droSec1.super_11 2329167 88 + 2888827 UGGCACACAUCAAAUGAAUUGUGUGCCGUGCAAUAAGGCAGAAAGGCAGCC---------CUCAGCACAAUGAGGAUUUUCACUUUUCUAGCUCACA .((((((((((....))..))))))))(((......((((((((((....(---------((((......))))).......))))))).)))))). ( -29.60, z-score = -2.91, R) >droYak2.chr3L 22821559 86 + 24197627 UGGCACACAUCAAAUGAAUUGUGUGCCGUACAAUAAGGAAGAAAG-CAGCC---------CUCAGCACAUUGAGGAUUU-CACUUUUCUAGCUCACA .((((((((((....))..)))))))).........((((((..(-.(..(---------(((((....))))))..).-)..))))))........ ( -23.30, z-score = -2.06, R) >droEre2.scaffold_4784 22062827 76 + 25762168 UGGCACACAUCAAAUGAGUUGUGUGCCGUACAAUAAGGCAGAAAGGCAGA---------------------GAGGCUUUUCACUUUUCUAGCUCACA .((((((((.(......).)))))))).........((((((((((..((---------------------((....)))).))))))).))).... ( -23.30, z-score = -2.24, R) >consensus UGGCACACAUCAAAUGAAUUGUGUGCCGUGCAAUAAGGCAGAAAGGCAGCC_________CUCAGCACAAUGAGGAUUUUCACUUUUCUAGCUCACA .((((((((((....))..)))))))).........((((((((((..............((((......))))(.....).))))))).))).... (-17.41 = -18.74 + 1.33)
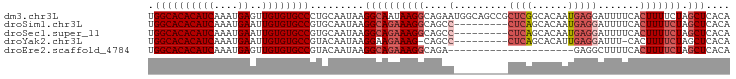
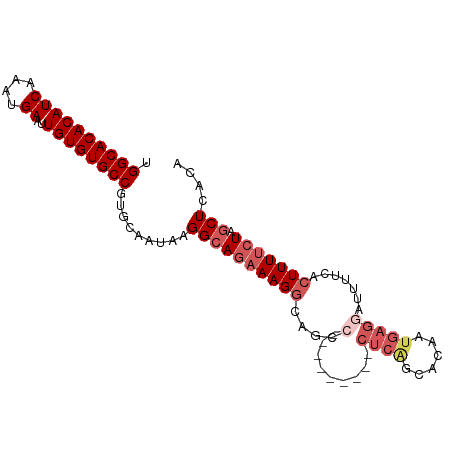
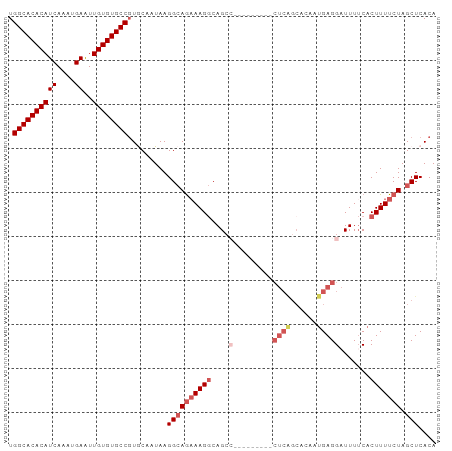
| Location | 22,392,618 – 22,392,715 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 97 |
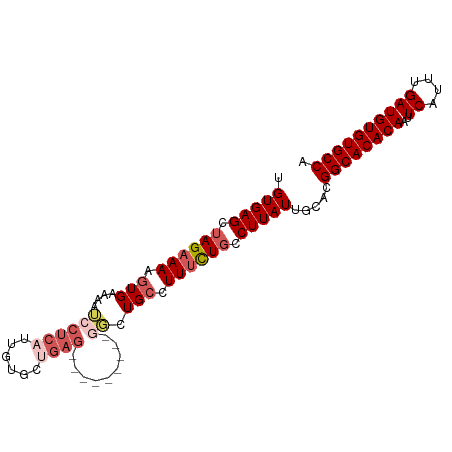
| Reading direction | reverse |
| Mean pairwise identity | 84.83 |
| Shannon entropy | 0.25849 |
| G+C content | 0.45218 |
| Mean single sequence MFE | -27.86 |
| Consensus MFE | -18.00 |
| Energy contribution | -18.44 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.40 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.02 |
| SVM RNA-class probability | 0.875359 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 22392618 97 - 24543557 UGUGAGCUAGAAAAGUGAAAAUCCUCAUUGUGCCGAGCGGCUGCCAUUCUGCCUUAUUGCCUUAUUGCAGGGCACACAACUCAUUUGAUGUGUGCCA .(((((.(((((.(((((......)))))..(((....))).....))))).)))))(((......))).((((((((.(......).)))))))). ( -29.30, z-score = -1.26, R) >droSim1.chr3L 21703816 88 - 22553184 UGUGAGCUAGAAAAGUGAAAAUCCUCAUUGUGCUGAG---------GGCUGCCUUUCUGCCUUAUUGCACGGCACACAAUUCAUUUGAUGUGUGCCA .............(((((......)))))((((..((---------(((.........)))))...))))((((((((..((....)))))))))). ( -29.50, z-score = -2.51, R) >droSec1.super_11 2329167 88 - 2888827 UGUGAGCUAGAAAAGUGAAAAUCCUCAUUGUGCUGAG---------GGCUGCCUUUCUGCCUUAUUGCACGGCACACAAUUCAUUUGAUGUGUGCCA .............(((((......)))))((((..((---------(((.........)))))...))))((((((((..((....)))))))))). ( -29.50, z-score = -2.51, R) >droYak2.chr3L 22821559 86 - 24197627 UGUGAGCUAGAAAAGUG-AAAUCCUCAAUGUGCUGAG---------GGCUG-CUUUCUUCCUUAUUGUACGGCACACAAUUCAUUUGAUGUGUGCCA .(((((..(((((.(((-...((((((......))))---------)).))-))))))..))))).....((((((((..((....)))))))))). ( -28.00, z-score = -2.81, R) >droEre2.scaffold_4784 22062827 76 - 25762168 UGUGAGCUAGAAAAGUGAAAAGCCUC---------------------UCUGCCUUUCUGCCUUAUUGUACGGCACACAACUCAUUUGAUGUGUGCCA .(((((.((((((.((((........---------------------)).)).)))))).))))).....((((((((.(......).)))))))). ( -23.00, z-score = -2.89, R) >consensus UGUGAGCUAGAAAAGUGAAAAUCCUCAUUGUGCUGAG_________GGCUGCCUUUCUGCCUUAUUGCACGGCACACAAUUCAUUUGAUGUGUGCCA .(((((.((((((.((..................................)).)))))).))))).....((((((((..((....)))))))))). (-18.00 = -18.44 + 0.44)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:44:40 2011