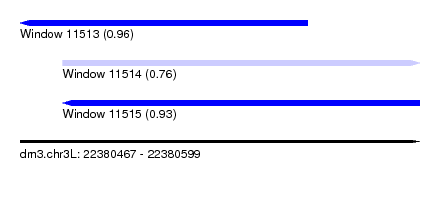
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 22,380,467 – 22,380,599 |
| Length | 132 |
| Max. P | 0.962941 |

| Location | 22,380,467 – 22,380,562 |
|---|---|
| Length | 95 |
| Sequences | 10 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 81.43 |
| Shannon entropy | 0.42338 |
| G+C content | 0.42253 |
| Mean single sequence MFE | -21.44 |
| Consensus MFE | -12.96 |
| Energy contribution | -13.21 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.71 |
| SVM RNA-class probability | 0.962941 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 22380467 95 - 24543557 AGCUAAUCAGCAAUAACUGUCAUUUAGCAAAUAGUUGCUCAUAUUUGCACUUAAGCAAAUAACCAGAGGAGGAG-CGGGGCACACUUGAGCUCCAC .(((((.(((......)))....)))))........((((.(((((((......))))))).((...))..)))-)(((((........))))).. ( -27.40, z-score = -2.57, R) >droSim1.chr3L_random 1000681 95 - 1049610 AGCUAAUCAGCAAUAACUGUCAUUUAGCAAAUAGUUGCUCAUAUUUGCACUUAAGCAAAUAACCAGAGAGGGAG-CGGUGCACACUCGAGCUCCAC .(((((.(((......)))....)))))........((((.(((((((......))))))).((.....)))))-)((.((........)).)).. ( -25.90, z-score = -2.20, R) >droSec1.super_11 2316828 95 - 2888827 AGCUAAUCAGCAAUAACUGUCAUUUAGCAAAUAGUUGCUCAUAUUUGCACUUAAGCAAAUAACCAGAGAGGGAG-CGGGGCACACUCGAGCUCCAC .(((((.(((......)))....)))))........((((.(((((((......))))))).((.....)))))-)(((((........))))).. ( -29.20, z-score = -3.47, R) >droYak2.chr3L 22807086 95 - 24197627 AGCUAAUCAGCAAUAACUGUCAUUUAGCAAAUAGUUGCUCAUAUUUGCACUUAAGCAAAUAACCAGAGGAGAAG-CGGGGCACACUCGGGUUCCAC .(((....))).....(((......(((((....)))))..(((((((......)))))))..))).((((...-((((.....))))..)))).. ( -22.70, z-score = -1.56, R) >droEre2.scaffold_4784 22050748 95 - 25762168 AGCUAAUCAGCAAUAACUGUCAUUUAGCAAAUAGUUGCUCAUAUUUGCACUUAAGCAAAUAACCACAGGAGAAG-CGGGGCACACACGAGCUCCAC .(((..((((((((...(((......)))....))))))..(((((((......))))))).......))..))-)(((((........))))).. ( -25.10, z-score = -2.93, R) >droAna3.scaffold_13337 22117317 93 - 23293914 AGCUAAUCAGCAAUAAUUGUCAUUUAGCAAAUAGUGGCUCAUACUUGCACUUAAGCAUACAGCCGGGCCCUU---CGAGGCGGACGCACACUCCCA .(((((...(((.....)))...)))))....((((((((.....(((......)))....(((........---...))).)).)).)))).... ( -19.20, z-score = 0.67, R) >dp4.chrXR_group8 33878 94 - 9212921 AGCUAAUCAGCAAUAACUGUCAUUUAGCAAAUAGUUGCUCAUAUUUGCACUUAAGCAAAUAACCAGAGACUC-CACUGUGCCC-CCCAAUCCCCCC .(((((.(((......)))....)))))..(((((.((((.(((((((......)))))))....))).)..-.)))))....-............ ( -17.30, z-score = -2.92, R) >droVir3.scaffold_13049 7838103 87 + 25233164 AGCUAAUCAGCAAUAACUGUCAUUUAGCAAAUAGUUGCUCAUAUUUGCACUUAAGCAAAUAACCAGGGACCGAGUCCUCACAUAUAC--------- .(((((.(((......)))....))))).............(((((((......)))))))....(((((...))))).........--------- ( -17.50, z-score = -1.96, R) >droMoj3.scaffold_6680 2321962 87 + 24764193 AGCUAAUCAGCAAUAACUGUCAUUUAGCAAAUAGUUGCUCAUAUUUGCACUUAAGCAAAUAACCAGAGACCGAAUCCCCGCACACAC--------- .(((((.(((......)))....))))).....(((((...(((((((......)))))))....((.......))...))).))..--------- ( -15.50, z-score = -2.27, R) >droGri2.scaffold_15110 14777749 76 - 24565398 AGCUAAUCAGCAAUAACUGUCAUUUAGCAAAUAGUUGCUCAUAUUUGCACUUAAGCAAAUAACCAGAGACCCAUAC-------------------- .(((((.(((......)))....))))).....((..((..(((((((......)))))))...))..))......-------------------- ( -14.60, z-score = -2.18, R) >consensus AGCUAAUCAGCAAUAACUGUCAUUUAGCAAAUAGUUGCUCAUAUUUGCACUUAAGCAAAUAACCAGAGACGGAG_CGGGGCACACUCGAGCUCCAC .(((((.(((......)))....))))).........(((.(((((((......)))))))....)))............................ (-12.96 = -13.21 + 0.25)
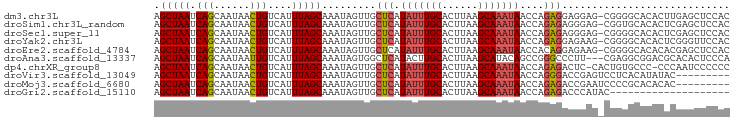
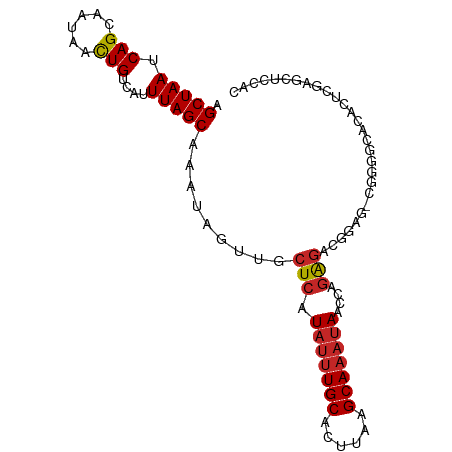
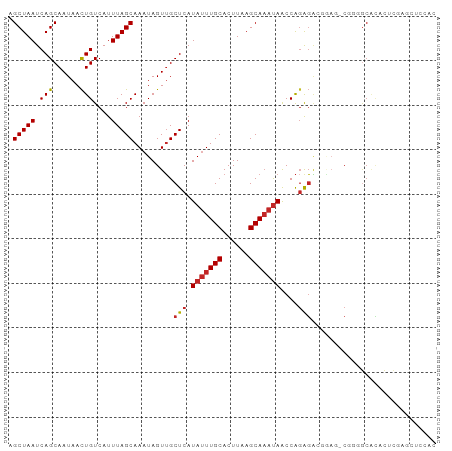
| Location | 22,380,481 – 22,380,599 |
|---|---|
| Length | 118 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.79 |
| Shannon entropy | 0.23663 |
| G+C content | 0.38655 |
| Mean single sequence MFE | -31.20 |
| Consensus MFE | -25.30 |
| Energy contribution | -25.83 |
| Covariance contribution | 0.52 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.81 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.758397 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 22380481 118 + 24543557 UGCCCCGCUCCUCCUCUGG-UUAUUUGCUUAAGUGCAAAUAUGAGCAACUAUUUGCUAAAUGACAGUUAUUGCUGAUUAGCUGUCAGACAUUUUGACAGC-AUGAAGAAUGAUUCGCAAU ......((((..((...))-.(((((((......))))))).))))......((((.......((((....))))....(((((((((...)))))))))-..(((......))))))). ( -30.30, z-score = -1.71, R) >droSim1.chr3L_random 1000695 118 + 1049610 UGCACCGCUCCCUCUCUGG-UUAUUUGCUUAAGUGCAAAUAUGAGCAACUAUUUGCUAAAUGACAGUUAUUGCUGAUUAGCUGUCAGACAUUUUGACAGC-AUGAAGAAUGAUUCGCAAU ......((((((.....))-.(((((((......))))))).))))......((((.......((((....))))....(((((((((...)))))))))-..(((......))))))). ( -32.60, z-score = -2.10, R) >droSec1.super_11 2316842 118 + 2888827 UGCCCCGCUCCCUCUCUGG-UUAUUUGCUUAAGUGCAAAUAUGAGCAACUAUUUGCUAAAUGACAGUUAUUGCUGAUUAGCUGUCAGACAUUUUGACAGC-AUGAAGAAUGAUUCGCAAU ......((((((.....))-.(((((((......))))))).))))......((((.......((((....))))....(((((((((...)))))))))-..(((......))))))). ( -32.60, z-score = -2.45, R) >droYak2.chr3L 22807100 117 + 24197627 UGCCCCGCUUCUCCUCUGG-UUAUUUGCUUAAGUGCAAAUAUGAGCAACUAUUUGCUAAAUGACAGUUAUUGCUGAUUAGCUGUCAGACAUUUUGACAGC-AUGAAGAAUGAUUCGCAU- (((....((((........-.(((((((......)))))))..(((((....)))))......((((....))))....(((((((((...)))))))))-..))))........))).- ( -28.90, z-score = -1.10, R) >droEre2.scaffold_4784 22050762 118 + 25762168 UGCCCCGCUUCUCCUGUGG-UUAUUUGCUUAAGUGCAAAUAUGAGCAACUAUUUGCUAAAUGACAGUUAUUGCUGAUUAGCUGUCAGACAUUUUGACAGC-AUGAAGAAUGAUUCGCAAU (((.((((.......))))-.(((((((......)))))))...))).....((((.......((((....))))....(((((((((...)))))))))-..(((......))))))). ( -32.30, z-score = -2.05, R) >droAna3.scaffold_13337 22117331 116 + 23293914 --CGCCUCGAAGGGCCCGG-CUGUAUGCUUAAGUGCAAGUAUGAGCCACUAUUUGCUAAAUGACAAUUAUUGCUGAUUAGCUGUCAGACGUUUUGACAGC-AUGAAGGAUGAUUCGCAAU --.((((....))))..((-((.(((((((......))))))))))).....................(((((......(((((((((...)))))))))-..(((......)))))))) ( -33.00, z-score = -1.04, R) >dp4.chrXR_group8 33891 118 + 9212921 GGCACAGUGGAGUCUCUGG-UUAUUUGCUUAAGUGCAAAUAUGAGCAACUAUUUGCUAAAUGACAGUUAUUGCUGAUUAGCUGUCAGACAUUUUGACAGC-UUGAAGAAUGAUUCGCAAU ((((.(((((...(((...-.(((((((......))))))).)))...)))))))))...((..(((((((.((....((((((((((...)))))))))-)...)))))))))..)).. ( -34.00, z-score = -1.71, R) >droPer1.super_80 47930 117 - 286535 GGGAUAGUGGAGUCUCUGG-UUAUUUGCUUAAGUGCAAAUAUGAGCAACUAUUUGCUAAAUGACAGUUAUUGCUGAUUAGCUGUCAGACAUUUU-ACAGC-UUGAAGAAUGAUUCGCAAU .(((((((...(.(((...-.(((((((......))))))).)))).)))))))((....(((((((((........)))))))))..((((((-.(...-..).))))))....))... ( -30.70, z-score = -1.28, R) >droWil1.scaffold_180698 3953713 120 - 11422946 UUCACUCUUCCACUUCUGGGUUAUUUGCUUAAGUGCAAAUAUGAGCAACUAUUUGCUAAAUGACAGUUAUUGCUGAUUAGCUGUCAGACAUUUUGACAGCCAUGAAGAAUGAUUCGCAAU .(((.(((((...........(((((((......)))))))..(((((....)))))..(((.((((....))))....(((((((((...))))))))))))))))).)))........ ( -32.00, z-score = -2.14, R) >droVir3.scaffold_13049 7838115 112 - 25233164 ------ACUCGGUCCCUGG-UUAUUUGCUUAAGUGCAAAUAUGAGCAACUAUUUGCUAAAUGACAGUUAUUGCUGAUUAGCUGUCAGGCAUUUUGACAGC-AUGAAGAAUGAUUCGCAAU ------.....(((.....-.(((((((......)))))))..(((((....)))))....)))....(((((......((((((((.....))))))))-..(((......)))))))) ( -28.80, z-score = -0.95, R) >droMoj3.scaffold_6680 2321974 111 - 24764193 ------AUUCGGUCUCUGG-UUAUUUGCUUAAGUGCAAAUAUGAGCAACUAUUUGCUAAAUGACAGUUAUUGCUGAUUAGCUGUCAGACAUUUUGACAGC-AUGAAGAAUGAUUCGCAA- ------..(((((..(((.-.(((((((......)))))))..(((((....)))))......))).....)))))...(((((((((...)))))))))-..(((......)))....- ( -29.30, z-score = -1.37, R) >droGri2.scaffold_15110 14777754 108 + 24565398 ----------GGUCUCUGG-UUAUUUGCUUAAGUGCAAAUAUGAGCAACUAUUUGCUAAAUGACAGUUAUUGCUGAUUAGCUGUCAGACAUUUUGACAGC-AUGAAGAAUGAUUCGCAAU ----------.(((.....-.(((((((......)))))))..(((((....)))))....)))....(((((......(((((((((...)))))))))-..(((......)))))))) ( -29.90, z-score = -1.81, R) >consensus UGCACCGCUCCGUCUCUGG_UUAUUUGCUUAAGUGCAAAUAUGAGCAACUAUUUGCUAAAUGACAGUUAUUGCUGAUUAGCUGUCAGACAUUUUGACAGC_AUGAAGAAUGAUUCGCAAU ...............(((...(((((((......)))))))..(((((....)))))......)))..(((((.((((((((((((((...))))))))).........))))).))))) (-25.30 = -25.83 + 0.52)
| Location | 22,380,481 – 22,380,599 |
|---|---|
| Length | 118 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.79 |
| Shannon entropy | 0.23663 |
| G+C content | 0.38655 |
| Mean single sequence MFE | -29.53 |
| Consensus MFE | -24.24 |
| Energy contribution | -24.84 |
| Covariance contribution | 0.61 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.82 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.33 |
| SVM RNA-class probability | 0.927138 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 22380481 118 - 24543557 AUUGCGAAUCAUUCUUCAU-GCUGUCAAAAUGUCUGACAGCUAAUCAGCAAUAACUGUCAUUUAGCAAAUAGUUGCUCAUAUUUGCACUUAAGCAAAUAA-CCAGAGGAGGAGCGGGGCA ..(((...((.((((((..-(((((((.......)))))))...((((((((...(((......)))....))))))..(((((((......))))))).-...)).)))))).)).))) ( -30.40, z-score = -1.24, R) >droSim1.chr3L_random 1000695 118 - 1049610 AUUGCGAAUCAUUCUUCAU-GCUGUCAAAAUGUCUGACAGCUAAUCAGCAAUAACUGUCAUUUAGCAAAUAGUUGCUCAUAUUUGCACUUAAGCAAAUAA-CCAGAGAGGGAGCGGUGCA ((((((((......)))..-(((((((.......)))))))......)))))............(((.....((((((.(((((((......))))))).-((.....))))))))))). ( -32.00, z-score = -1.69, R) >droSec1.super_11 2316842 118 - 2888827 AUUGCGAAUCAUUCUUCAU-GCUGUCAAAAUGUCUGACAGCUAAUCAGCAAUAACUGUCAUUUAGCAAAUAGUUGCUCAUAUUUGCACUUAAGCAAAUAA-CCAGAGAGGGAGCGGGGCA ((((((((......)))..-(((((((.......)))))))......)))))....(((....(((.....)))((((.(((((((......))))))).-((.....))))))..))). ( -31.60, z-score = -1.63, R) >droYak2.chr3L 22807100 117 - 24197627 -AUGCGAAUCAUUCUUCAU-GCUGUCAAAAUGUCUGACAGCUAAUCAGCAAUAACUGUCAUUUAGCAAAUAGUUGCUCAUAUUUGCACUUAAGCAAAUAA-CCAGAGGAGAAGCGGGGCA -.(((...((.((((((..-(((((((.......)))))))...((((((((...(((......)))....))))))..(((((((......))))))).-...))))))))..)).))) ( -30.50, z-score = -1.55, R) >droEre2.scaffold_4784 22050762 118 - 25762168 AUUGCGAAUCAUUCUUCAU-GCUGUCAAAAUGUCUGACAGCUAAUCAGCAAUAACUGUCAUUUAGCAAAUAGUUGCUCAUAUUUGCACUUAAGCAAAUAA-CCACAGGAGAAGCGGGGCA ..(((...((.((((((..-(((((((.......))))))).....((((((...(((......)))....))))))..(((((((......))))))).-.....))))))..)).))) ( -29.70, z-score = -1.56, R) >droAna3.scaffold_13337 22117331 116 - 23293914 AUUGCGAAUCAUCCUUCAU-GCUGUCAAAACGUCUGACAGCUAAUCAGCAAUAAUUGUCAUUUAGCAAAUAGUGGCUCAUACUUGCACUUAAGCAUACAG-CCGGGCCCUUCGAGGCG-- ((((((((......)))..-(((((((.......)))))))......)))))............((....((.(((((.....(((......))).....-..))))))).....)).-- ( -28.60, z-score = -0.97, R) >dp4.chrXR_group8 33891 118 - 9212921 AUUGCGAAUCAUUCUUCAA-GCUGUCAAAAUGUCUGACAGCUAAUCAGCAAUAACUGUCAUUUAGCAAAUAGUUGCUCAUAUUUGCACUUAAGCAAAUAA-CCAGAGACUCCACUGUGCC ((((((((......))).(-(((((((.......)))))))).....)))))....(((....(((((....)))))..(((((((......))))))).-.....)))........... ( -29.20, z-score = -2.52, R) >droPer1.super_80 47930 117 + 286535 AUUGCGAAUCAUUCUUCAA-GCUGU-AAAAUGUCUGACAGCUAAUCAGCAAUAACUGUCAUUUAGCAAAUAGUUGCUCAUAUUUGCACUUAAGCAAAUAA-CCAGAGACUCCACUAUCCC ((((((((......))).(-(((((-..........)))))).....)))))....(((....(((((....)))))..(((((((......))))))).-.....)))........... ( -24.20, z-score = -2.17, R) >droWil1.scaffold_180698 3953713 120 + 11422946 AUUGCGAAUCAUUCUUCAUGGCUGUCAAAAUGUCUGACAGCUAAUCAGCAAUAACUGUCAUUUAGCAAAUAGUUGCUCAUAUUUGCACUUAAGCAAAUAACCCAGAAGUGGAAGAGUGAA ........(((((((((.(((((((((.......)))))))))..(((......))).((((((((((....)))))..(((((((......)))))))......)))))))))))))). ( -36.60, z-score = -3.70, R) >droVir3.scaffold_13049 7838115 112 + 25233164 AUUGCGAAUCAUUCUUCAU-GCUGUCAAAAUGCCUGACAGCUAAUCAGCAAUAACUGUCAUUUAGCAAAUAGUUGCUCAUAUUUGCACUUAAGCAAAUAA-CCAGGGACCGAGU------ ((((((((......)))..-(((((((.......)))))))......)))))..(.(((....(((((....)))))..(((((((......))))))).-.....))).)...------ ( -27.60, z-score = -1.93, R) >droMoj3.scaffold_6680 2321974 111 + 24764193 -UUGCGAAUCAUUCUUCAU-GCUGUCAAAAUGUCUGACAGCUAAUCAGCAAUAACUGUCAUUUAGCAAAUAGUUGCUCAUAUUUGCACUUAAGCAAAUAA-CCAGAGACCGAAU------ -(((((((......)))..-(((((((.......)))))))......))))...(.(((....(((((....)))))..(((((((......))))))).-.....))).)...------ ( -26.60, z-score = -2.39, R) >droGri2.scaffold_15110 14777754 108 - 24565398 AUUGCGAAUCAUUCUUCAU-GCUGUCAAAAUGUCUGACAGCUAAUCAGCAAUAACUGUCAUUUAGCAAAUAGUUGCUCAUAUUUGCACUUAAGCAAAUAA-CCAGAGACC---------- ((((((((......)))..-(((((((.......)))))))......)))))....(((....(((((....)))))..(((((((......))))))).-.....))).---------- ( -27.40, z-score = -2.93, R) >consensus AUUGCGAAUCAUUCUUCAU_GCUGUCAAAAUGUCUGACAGCUAAUCAGCAAUAACUGUCAUUUAGCAAAUAGUUGCUCAUAUUUGCACUUAAGCAAAUAA_CCAGAGACGGAGCGGGGCA ((((((((......)))...(((((((.......)))))))......)))))..(((......(((((....)))))..(((((((......)))))))...)))............... (-24.24 = -24.84 + 0.61)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:44:36 2011