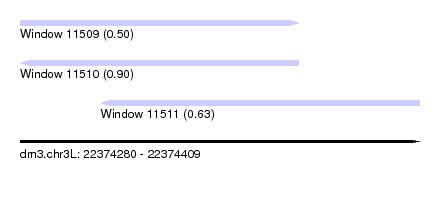
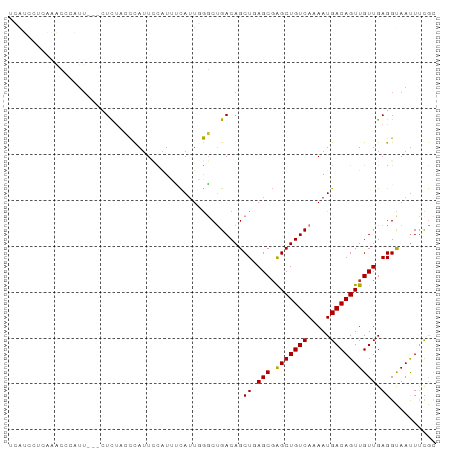
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 22,374,280 – 22,374,409 |
| Length | 129 |
| Max. P | 0.899757 |

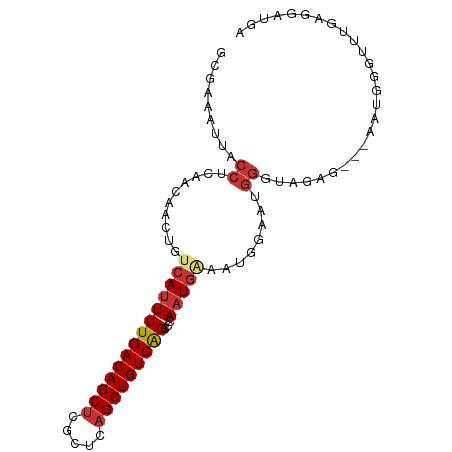
| Location | 22,374,280 – 22,374,370 |
|---|---|
| Length | 90 |
| Sequences | 8 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 76.64 |
| Shannon entropy | 0.47305 |
| G+C content | 0.47403 |
| Mean single sequence MFE | -25.92 |
| Consensus MFE | -11.92 |
| Energy contribution | -12.53 |
| Covariance contribution | 0.61 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
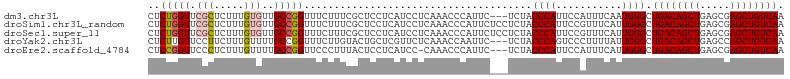
>dm3.chr3L 22374280 90 + 24543557 ACGAAGUUACCUCAACAACUGUCAUUUUGACAGCUCGCUCAGCUGUCAGCCCAUUGAAAUGGAAUGGGUAGAG---AAUGGGUUUGAGGAUGA .........((((((..(((..(((((((((((((.....))))))).(((((((.......)))))))..))---))))))))))))).... ( -30.60, z-score = -2.58, R) >droSim1.chr3L_random 993954 93 + 1049610 GCGAAAUUACCUCAACAACUGUCAUUUUGACAGCUCGCUCAGCUGUCAGCCCAAUGAAACGGAAUGGGUAGAGGAGAAUGGGUUUGAGGAUGA .........((((((..(((..(((((((((((((.....))))))).(((((...........))))).....))))))))))))))).... ( -27.50, z-score = -1.40, R) >droSec1.super_11 2310613 93 + 2888827 GCGAAAUUACCUCAACAACUGUCAUUUUGACAGCUCGCUCAGCUGUCAGCCCAAUGAAACGGAAUGGGUAGAGGAGAAUGGGUUUGAGGAUGA .........((((((..(((..(((((((((((((.....))))))).(((((...........))))).....))))))))))))))).... ( -27.50, z-score = -1.40, R) >droYak2.chr3L 22801024 90 + 24197627 GUGAAAUUGCCUCAACAACUGUCAUUUUGACAGCUGGCUCAGCUGUCAGCCCAAUAAAAGGGACUGGGUAGAG---AAUUGGUUUGAGAACGA ..........((((.(((.(.((....(((((((((...)))))))))(((((...........))))).)).---).)))...))))..... ( -26.00, z-score = -1.12, R) >droEre2.scaffold_4784 22044510 89 + 25762168 GCGAAAUUACCUCAACAACUGUCAUUUUGACAGCUCGCUCAGCUGUCAGCCCAAUGAAAUGGAAUGGGUAGAG---AAUGGGUUUG-GGAUGA .........(((.(((.....((....((((((((.....))))))))(((((...........)))))...)---)....))).)-)).... ( -23.50, z-score = -0.44, R) >dp4.chrXR_group8 25261 78 + 9212921 GCGGAAUUGCCUCAACAACUGUCAUUUUGACAGCUCGCUCAGCUGUCGGGCGAAUGCCAUGUAAUGGCGGAAGCACAA--------------- ((((.....)).......((((((((.((((((((.....))))))))(((....)))....))))))))..))....--------------- ( -25.90, z-score = -1.36, R) >droPer1.super_80 39285 76 - 286535 GCGGAAUUGCCUCAACAACUGUCAUUUUGACAGCUCGCUCAGCUGUCGGGCGAAUGCCAUGUAAUGGCGGAAACAC----------------- (.((.....)).).....((((((((.((((((((.....))))))))(((....)))....))))))))......----------------- ( -23.70, z-score = -1.41, R) >droWil1.scaffold_180698 3945173 90 - 11422946 GCAAAAUUACCUCAACAACUGUCAUUUUGACAGCCCGCUCAGCUGUCAGCUGAAUGGAACAAAAUAACAAAAC---AAGAUGCCAGAGCAUCA ..................(((((.....))))).(((.(((((.....))))).)))................---..(((((....))))). ( -22.70, z-score = -2.96, R) >consensus GCGAAAUUACCUCAACAACUGUCAUUUUGACAGCUCGCUCAGCUGUCAGCCCAAUGAAAUGGAAUGGGUAGAG___AAUGGGUUUGAGGAUGA .........((..........((((((((((((((.....)))))))))...)))))........)).......................... (-11.92 = -12.53 + 0.61)

| Location | 22,374,280 – 22,374,370 |
|---|---|
| Length | 90 |
| Sequences | 8 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 76.64 |
| Shannon entropy | 0.47305 |
| G+C content | 0.47403 |
| Mean single sequence MFE | -24.60 |
| Consensus MFE | -13.51 |
| Energy contribution | -13.40 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.15 |
| SVM RNA-class probability | 0.899757 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
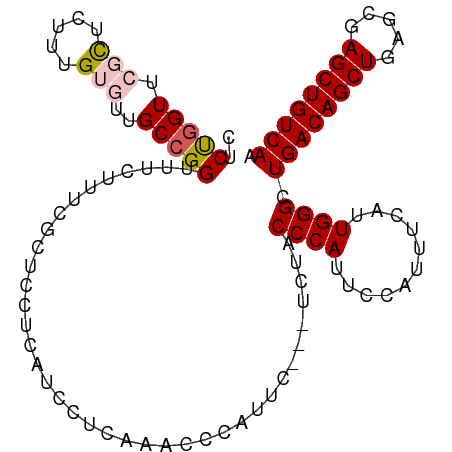
>dm3.chr3L 22374280 90 - 24543557 UCAUCCUCAAACCCAUU---CUCUACCCAUUCCAUUUCAAUGGGCUGACAGCUGAGCGAGCUGUCAAAAUGACAGUUGUUGAGGUAACUUCGU ....((((((....(((---.((..((((((.......)))))).((((((((.....))))))))....)).)))..))))))......... ( -25.20, z-score = -2.31, R) >droSim1.chr3L_random 993954 93 - 1049610 UCAUCCUCAAACCCAUUCUCCUCUACCCAUUCCGUUUCAUUGGGCUGACAGCUGAGCGAGCUGUCAAAAUGACAGUUGUUGAGGUAAUUUCGC ....((((((......((.......((((...........)))).((((((((.....))))))))....))......))))))......... ( -23.50, z-score = -1.52, R) >droSec1.super_11 2310613 93 - 2888827 UCAUCCUCAAACCCAUUCUCCUCUACCCAUUCCGUUUCAUUGGGCUGACAGCUGAGCGAGCUGUCAAAAUGACAGUUGUUGAGGUAAUUUCGC ....((((((......((.......((((...........)))).((((((((.....))))))))....))......))))))......... ( -23.50, z-score = -1.52, R) >droYak2.chr3L 22801024 90 - 24197627 UCGUUCUCAAACCAAUU---CUCUACCCAGUCCCUUUUAUUGGGCUGACAGCUGAGCCAGCUGUCAAAAUGACAGUUGUUGAGGCAAUUUCAC ..((.(((((..(((((---.((..((((((.......)))))).(((((((((...)))))))))....)).))))))))))))........ ( -27.60, z-score = -2.98, R) >droEre2.scaffold_4784 22044510 89 - 25762168 UCAUCC-CAAACCCAUU---CUCUACCCAUUCCAUUUCAUUGGGCUGACAGCUGAGCGAGCUGUCAAAAUGACAGUUGUUGAGGUAAUUUCGC ......-...(((((..---..((.((((...........)))).((((((((.....)))))))).......))....)).)))........ ( -20.00, z-score = -0.70, R) >dp4.chrXR_group8 25261 78 - 9212921 ---------------UUGUGCUUCCGCCAUUACAUGGCAUUCGCCCGACAGCUGAGCGAGCUGUCAAAAUGACAGUUGUUGAGGCAAUUCCGC ---------------...((((((.(((((...)))))..((((.((.....)).))))((((((.....))))))....))))))....... ( -24.70, z-score = -1.37, R) >droPer1.super_80 39285 76 + 286535 -----------------GUGUUUCCGCCAUUACAUGGCAUUCGCCCGACAGCUGAGCGAGCUGUCAAAAUGACAGUUGUUGAGGCAAUUCCGC -----------------(((.....(((((...)))))....(((((((.((...)).(((((((.....))))))))))).))).....))) ( -24.40, z-score = -1.67, R) >droWil1.scaffold_180698 3945173 90 + 11422946 UGAUGCUCUGGCAUCUU---GUUUUGUUAUUUUGUUCCAUUCAGCUGACAGCUGAGCGGGCUGUCAAAAUGACAGUUGUUGAGGUAAUUUUGC .(((((....)))))..---((...((((((((...((.((((((.....)))))).))((((((.....))))))....))))))))...)) ( -27.90, z-score = -2.00, R) >consensus UCAUCCUCAAACCCAUU___CUCUACCCAUUCCAUUUCAUUGGGCUGACAGCUGAGCGAGCUGUCAAAAUGACAGUUGUUGAGGUAAUUUCGC ...................................................((.(((.(((((((.....)))))))))).)).......... (-13.51 = -13.40 + -0.11)

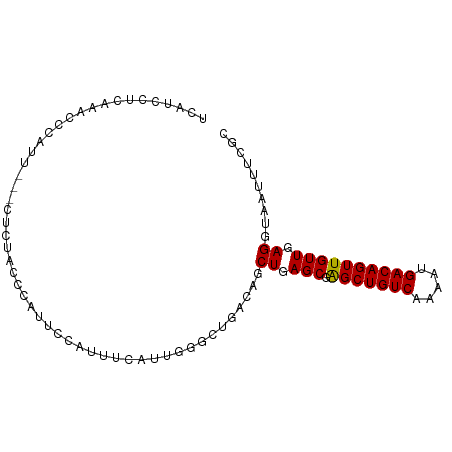
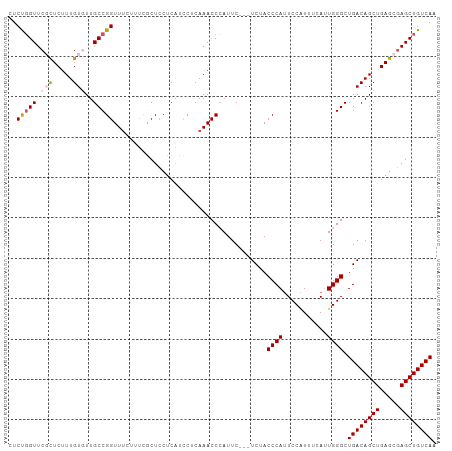
| Location | 22,374,306 – 22,374,409 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 89.53 |
| Shannon entropy | 0.18064 |
| G+C content | 0.50368 |
| Mean single sequence MFE | -23.91 |
| Consensus MFE | -19.60 |
| Energy contribution | -20.28 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.82 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.630571 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 22374306 103 - 24543557 CUCUGGUUCGCUCUUUGUGUUGCCGGUUUCUUUCGCUCCUCAUCCUCAAACCCAUUC---UCUACCCAUUCCAUUUCAAUGGGCUGACAGCUGAGCGAGCUGUCAA ..(((((.(((.....)))..)))))...............................---....((((((.......)))))).((((((((.....)))))))). ( -26.00, z-score = -1.99, R) >droSim1.chr3L_random 993980 106 - 1049610 CUCUGGUUCGCUCUUUGUGUUGCCGGUUUCUUUCGCUCCUCAUCCUCAAACCCAUUCUCCUCUACCCAUUCCGUUUCAUUGGGCUGACAGCUGAGCGAGCUGUCAA ..(((((.(((.....)))..)))))......................................((((...........)))).((((((((.....)))))))). ( -24.30, z-score = -1.25, R) >droSec1.super_11 2310639 106 - 2888827 CUCUGGUUCGCUCUUUGUGUUGCCGGUUUCUUUCGCUCCUCAUCCUCAAACCCAUUCUCCUCUACCCAUUCCGUUUCAUUGGGCUGACAGCUGAGCGAGCUGUCAA ..(((((.(((.....)))..)))))......................................((((...........)))).((((((((.....)))))))). ( -24.30, z-score = -1.25, R) >droYak2.chr3L 22801050 103 - 24197627 CUCUUGUUCCUUCUUUGUUUUGCCGGUUUCUUGUACUGCUCGUUCUCAAACCAAUUC---UCUACCCAGUCCCUUUUAUUGGGCUGACAGCUGAGCCAGCUGUCAA ........................(((((...(.((.....)).)..))))).....---....((((((.......)))))).(((((((((...))))))))). ( -22.80, z-score = -1.85, R) >droEre2.scaffold_4784 22044536 102 - 25762168 CUCCGGUUCCCUCUUUGUUUUGCCGGUUCCCUUUACUCCUCAUCC-CAAACCCAUUC---UCUACCCAUUCCAUUUCAUUGGGCUGACAGCUGAGCGAGCUGUCAA ..(((((..............)))))...................-...........---....((((...........)))).((((((((.....)))))))). ( -22.14, z-score = -1.84, R) >consensus CUCUGGUUCGCUCUUUGUGUUGCCGGUUUCUUUCGCUCCUCAUCCUCAAACCCAUUC___UCUACCCAUUCCAUUUCAUUGGGCUGACAGCUGAGCGAGCUGUCAA ..(((((.(((.....)))..)))))......................................((((...........)))).((((((((.....)))))))). (-19.60 = -20.28 + 0.68)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:44:33 2011