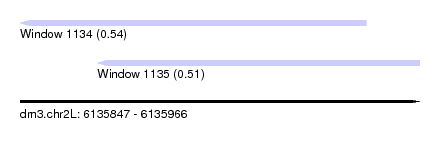
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 6,135,847 – 6,135,966 |
| Length | 119 |
| Max. P | 0.536690 |

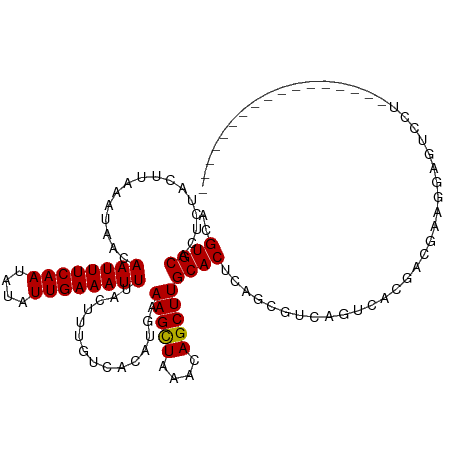
| Location | 6,135,847 – 6,135,950 |
|---|---|
| Length | 103 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.45 |
| Shannon entropy | 0.34360 |
| G+C content | 0.37594 |
| Mean single sequence MFE | -19.77 |
| Consensus MFE | -13.65 |
| Energy contribution | -13.57 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.37 |
| Structure conservation index | 0.69 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.536690 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 6135847 103 - 23011544 ACGUGCACUCUACUUAAAUAACAAUUUCAAUAUAUUGAAAUUUACUUUGUCACAUGAAAGCUAAACAGCUUGCACUCAGCGUCAGUCACGACAAAGGAGUCCU----------------- ....(.((((............((((((((....))))))))..(((((((...((((((((....)))))((.....)).))).....))))))))))).).----------------- ( -21.80, z-score = -2.23, R) >droGri2.scaffold_15252 14444283 118 + 17193109 ACGUGCACUCUACUUAAAUAACAAUUUCAAUAUAUUGAAAUUUACUUUGUCACAUGAAAGCUAAACAGCUUGCACUCAGCGUCAGUCACGAAGCAAUACACUUGGCUGGCCUACCCCC-- ..((((................((((((((....))))))))...............(((((....)))))))))..((.((((((((.(..........).))))))))))......-- ( -23.70, z-score = -1.71, R) >droMoj3.scaffold_6500 18853772 120 + 32352404 ACGUGCACUCUACUUAAAUAACAAUUUCAAUAUAUUGAAAUUUACUUUGUCACAUGAAAGCUAAACAGCUUGCACUCAGCGUCAGUCACGAAGCAAUACACAAGACUUGCCCAGCUCCAA ..(.(((.(((...........((((((((....))))))))..((((((.((.((((((((....)))))((.....)).))))).)))))).........)))..))).)........ ( -20.20, z-score = -0.81, R) >droVir3.scaffold_12963 17057580 120 + 20206255 ACGUGCACUCUACUUAAAUAACAAUUUCAAUAUAUUGAAAUUUACUUUGUCACAUGAAAGCUAAACAGCUUGCACUCAGCGUCAGUCACGAAGCAAUCCACAAGCUACACCCGCCACACA ..((((................((((((((....))))))))..((((((.((.((((((((....)))))((.....)).))))).))))))...................).)))... ( -19.80, z-score = -1.45, R) >droWil1.scaffold_180708 4913269 111 + 12563649 AUGUGCACUCUACUUAAAUAACAAUUUCAAUAUAUUGAAAUUUACUUUGUCACAUGAAAGCUAAACAGCUUGCACUCAGCGUCAGUCACAACGAAGGAGUAUAACAACGCA--------- .(((..(((((...........((((((((....))))))))..(.((((.((.((((((((....)))))((.....)).))))).)))).)..)))))...))).....--------- ( -20.40, z-score = -0.98, R) >droPer1.super_1 10234989 109 + 10282868 ACGUGCACUCUACUUAAAUAACAAUUUCAAUAUAUUGAAAUUUACUUUGUCACAUGAAAGCUAAACAGCUUGCACUCAGCGUCAGUCACAGCGAAGGAGUCCCAUCCCG----------- ..(.(.((((............((((((((....))))))))..((((((....((((((((....)))))((.....)).)))......)))))))))).))......----------- ( -18.80, z-score = -0.83, R) >dp4.chr4_group1 5230132 109 + 5278887 ACGUGCACUCUACUUAAAUAACAAUUUCAAUAUAUUGAAAUUUACUUUGUCACAUGAAAGCUAAACAGCUUGCACUCAGCGUCAGUCACAGCGAAGGAGUCCCAUCCCG----------- ..(.(.((((............((((((((....))))))))..((((((....((((((((....)))))((.....)).)))......)))))))))).))......----------- ( -18.80, z-score = -0.83, R) >droAna3.scaffold_12916 8124107 103 + 16180835 ACGUGCACUCUACUUAAAUAACAAUUUCAAUAUAUUGAAAUUUACUUUGUCACAUGAAAGUUAAACAGCUUGCACUUAGCUUCAGUCACGUCGAAGGUUCCCU----------------- ..(((((..((...........((((((((....)))))))).(((((........))))).....))..)))))....((((.........)))).......----------------- ( -14.50, z-score = -0.07, R) >droEre2.scaffold_4929 15060010 103 - 26641161 ACGUGCACUCUACUUAAAUAACAAUUUCAAUAUAUUGAAAUUUACUUUGUCACAUGAAAGCUAAACAGCUUGCACUCAGCGUCAGUCACGACAAAGGAGUCCU----------------- ....(.((((............((((((((....))))))))..(((((((...((((((((....)))))((.....)).))).....))))))))))).).----------------- ( -21.80, z-score = -2.23, R) >droYak2.chr2L 15546473 103 + 22324452 ACGUGCACUCUACUUAAAUAACAAUUUCAAUAUAUUGAAAUUUACUUUGUCACAUGAAAGCUAAACAGCUUGCACUCAGCGUCAGUCACGACAAAGGAGUCCU----------------- ....(.((((............((((((((....))))))))..(((((((...((((((((....)))))((.....)).))).....))))))))))).).----------------- ( -21.80, z-score = -2.23, R) >droSec1.super_5 4206007 86 - 5866729 ACGUGCACUCUACUUAAAUAACAAUUUCAAUAUAUUGAAAUUUACUUUGUCACAUGAAAGCUAAACAGCUUGCACUCAGCGUCAGA---------------------------------- ..((((................((((((((....))))))))...............(((((....)))))))))...........---------------------------------- ( -13.80, z-score = -0.89, R) >droSim1.chr2L 5923339 103 - 22036055 ACGUGCACUCUACUUAAAUAACAAUUUCAAUAUAUUGAAAUUUACUUUGUCACAUGAAAGCUAAACAGCUUGCACUCAGCGUCAGUCACGACAAAGGAGUCCU----------------- ....(.((((............((((((((....))))))))..(((((((...((((((((....)))))((.....)).))).....))))))))))).).----------------- ( -21.80, z-score = -2.23, R) >consensus ACGUGCACUCUACUUAAAUAACAAUUUCAAUAUAUUGAAAUUUACUUUGUCACAUGAAAGCUAAACAGCUUGCACUCAGCGUCAGUCACGACGAAGGAGUCCU_________________ ..((((................((((((((....))))))))...............(((((....)))))))))............................................. (-13.65 = -13.57 + -0.08)

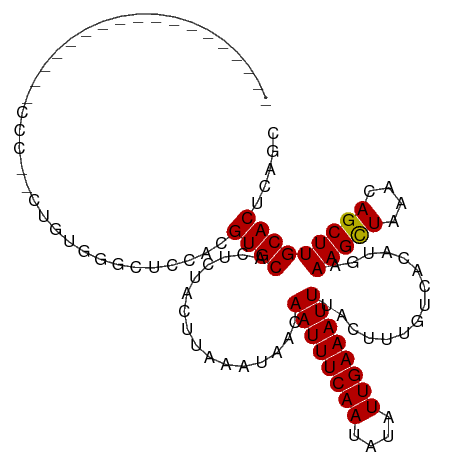
| Location | 6,135,870 – 6,135,966 |
|---|---|
| Length | 96 |
| Sequences | 11 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 87.10 |
| Shannon entropy | 0.27363 |
| G+C content | 0.38426 |
| Mean single sequence MFE | -18.54 |
| Consensus MFE | -13.64 |
| Energy contribution | -13.55 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.74 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.507239 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 6135870 96 - 23011544 ------------------CACCCCUGUGGGCUCCACGUGCACUCUACUUAAAUAACAAUUUCAAUAUAUUGAAAUUUACUUUGUCACAUGAAAGCUAAACAGCUUGCACUCAGC ------------------.......((((...))))((((................((((((((....))))))))...............(((((....)))))))))..... ( -17.40, z-score = -1.34, R) >droGri2.scaffold_15252 14444321 93 + 17193109 ---------------------CACCACUGGCUCCACGUGCACUCUACUUAAAUAACAAUUUCAAUAUAUUGAAAUUUACUUUGUCACAUGAAAGCUAAACAGCUUGCACUCAGC ---------------------........(((....((((................((((((((....))))))))...............(((((....)))))))))..))) ( -14.10, z-score = -1.11, R) >droMoj3.scaffold_6500 18853812 104 + 32352404 ----------CACCUACAUCUGCUGGCUGGUUCCACGUGCACUCUACUUAAAUAACAAUUUCAAUAUAUUGAAAUUUACUUUGUCACAUGAAAGCUAAACAGCUUGCACUCAGC ----------...........(((((.((....)).((((................((((((((....))))))))...............(((((....)))))))))))))) ( -17.90, z-score = -0.96, R) >droVir3.scaffold_12963 17057620 86 + 20206255 ----------------------------GGCUCCACGUGCACUCUACUUAAAUAACAAUUUCAAUAUAUUGAAAUUUACUUUGUCACAUGAAAGCUAAACAGCUUGCACUCAGC ----------------------------.(((....((((................((((((((....))))))))...............(((((....)))))))))..))) ( -14.10, z-score = -1.41, R) >droWil1.scaffold_180708 4913300 89 + 12563649 -------------------------GAGGUCUCCAUGUGCACUCUACUUAAAUAACAAUUUCAAUAUAUUGAAAUUUACUUUGUCACAUGAAAGCUAAACAGCUUGCACUCAGC -------------------------(((((...((((((.((......(((((..((((........))))..)))))....)))))))).(((((....))))))).)))... ( -16.80, z-score = -1.75, R) >droPer1.super_1 10235018 114 + 10282868 CAUCACUGGGUGGGUGUGGGUGUGGGUGGGAUCCACGUGCACUCUACUUAAAUAACAAUUUCAAUAUAUUGAAAUUUACUUUGUCACAUGAAAGCUAAACAGCUUGCACUCAGC .....(((((((.(((((((((((.((((...)))).)))))).............((((((((....))))))))........)))))..(((((....))))).))))))). ( -34.10, z-score = -2.65, R) >droAna3.scaffold_12916 8124130 98 + 16180835 ----------------CACCCAUUGGGAGGCUCCACGUGCACUCUACUUAAAUAACAAUUUCAAUAUAUUGAAAUUUACUUUGUCACAUGAAAGUUAAACAGCUUGCACUUAGC ----------------..(((...)))..(((....(((((..((...........((((((((....)))))))).(((((........))))).....))..)))))..))) ( -15.30, z-score = -0.24, R) >droEre2.scaffold_4929 15060033 96 - 26641161 ----------------CACCC--UUGUGGGCUCCACGUGCACUCUACUUAAAUAACAAUUUCAAUAUAUUGAAAUUUACUUUGUCACAUGAAAGCUAAACAGCUUGCACUCAGC ----------------..(((--....)))......((((................((((((((....))))))))...............(((((....)))))))))..... ( -17.70, z-score = -1.45, R) >droYak2.chr2L 15546496 96 + 22324452 ----------------CGCCC--UUGUGGGCUCCACGUGCACUCUACUUAAAUAACAAUUUCAAUAUAUUGAAAUUUACUUUGUCACAUGAAAGCUAAACAGCUUGCACUCAGC ----------------.((((--....)))).....((((................((((((((....))))))))...............(((((....)))))))))..... ( -21.70, z-score = -2.56, R) >droSec1.super_5 4206013 96 - 5866729 ------------------CACCCCUGUGGGCUCCACGUGCACUCUACUUAAAUAACAAUUUCAAUAUAUUGAAAUUUACUUUGUCACAUGAAAGCUAAACAGCUUGCACUCAGC ------------------.......((((...))))((((................((((((((....))))))))...............(((((....)))))))))..... ( -17.40, z-score = -1.34, R) >droSim1.chr2L 5923362 96 - 22036055 ------------------CACCCCUGUGGGCUCCACGUGCACUCUACUUAAAUAACAAUUUCAAUAUAUUGAAAUUUACUUUGUCACAUGAAAGCUAAACAGCUUGCACUCAGC ------------------.......((((...))))((((................((((((((....))))))))...............(((((....)))))))))..... ( -17.40, z-score = -1.34, R) >consensus __________________CCC__CUGUGGGCUCCACGUGCACUCUACUUAAAUAACAAUUUCAAUAUAUUGAAAUUUACUUUGUCACAUGAAAGCUAAACAGCUUGCACUCAGC ....................................((((................((((((((....))))))))...............(((((....)))))))))..... (-13.64 = -13.55 + -0.08)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:20:28 2011