
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 22,334,221 – 22,334,345 |
| Length | 124 |
| Max. P | 0.886086 |

| Location | 22,334,221 – 22,334,311 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 90 |
| Reading direction | reverse |
| Mean pairwise identity | 95.78 |
| Shannon entropy | 0.07247 |
| G+C content | 0.37778 |
| Mean single sequence MFE | -15.56 |
| Consensus MFE | -14.56 |
| Energy contribution | -14.76 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.94 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.07 |
| SVM RNA-class probability | 0.886086 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
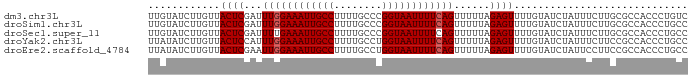
>dm3.chr3L 22334221 90 - 24543557 UUGUAUCUUGUUACUCGAUUUGGAAAUUGCCUUUUGCCCGGUAAUUUUCAGUUUUUAGAGUUUUGUAUCUAUUUCUUGCGCCACCCUGUC ............((((...((((((((((((........))))))))))))......))))............................. ( -15.60, z-score = -1.43, R) >droSim1.chr3L 21668570 90 - 22553184 UUGUAUCUUGUUACUCGAUUUGGAAAUUGCCUUUUGCCCGGUAAUUUUCAGUUUUUAGAGUUUUGUAUCUAUUUCUUGCGCCACCCUGCC ............((((...((((((((((((........))))))))))))......))))................(((......))). ( -16.00, z-score = -1.43, R) >droSec1.super_11 2270266 90 - 2888827 UUGUAUCUUGUUACUCGAUUUUGAAAUUGCCUUUUGCCCGGUAAUUUUCAGUUUUUAGAGUUUUGUAUCUAUUUCUUGCGCCACCCUGCC ............((((....(((((((((((........))))).))))))......))))................(((......))). ( -12.20, z-score = -0.68, R) >droYak2.chr3L 22747099 90 - 24197627 UUAUAUCUUGUUACUCCAUUUGGAAAUUGCCUUUUGCCUGGUAAUUUUCAGUUUUUAGAGUUUUGUAUCUAUUUCUUCCGCCACCCUGCC ............((((...((((((((((((........))))))))))))......))))............................. ( -15.10, z-score = -2.03, R) >droEre2.scaffold_4784 22004590 90 - 25762168 UUAUAUCUUGUUACUCGAAUUGGAAAUUGCCUUUUGCCUGGUAAUUUUCAGUUUUUAGAGUUUUGUAUCUAUUCCUUCCGCCACCCUGCC ............(((((((((((((((((((........)))))))))))))))...))))............................. ( -18.90, z-score = -3.19, R) >consensus UUGUAUCUUGUUACUCGAUUUGGAAAUUGCCUUUUGCCCGGUAAUUUUCAGUUUUUAGAGUUUUGUAUCUAUUUCUUGCGCCACCCUGCC ............((((...((((((((((((........))))))))))))......))))............................. (-14.56 = -14.76 + 0.20)


| Location | 22,334,245 – 22,334,345 |
|---|---|
| Length | 100 |
| Sequences | 7 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 82.00 |
| Shannon entropy | 0.35010 |
| G+C content | 0.29638 |
| Mean single sequence MFE | -16.76 |
| Consensus MFE | -10.89 |
| Energy contribution | -12.20 |
| Covariance contribution | 1.31 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.750176 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
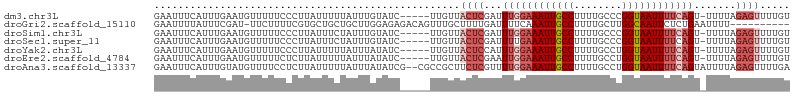
>dm3.chr3L 22334245 100 - 24543557 GAAUUUCAUUUGAAUGUUUUUCCCUUAUUUUUAUUUGUAUC-----UUGUUACUCGAUUUGGAAAUUGCCUUUUGCCCGGUAAUUUUCAGU-UUUUAGAGUUUUGU (((...(((....)))...)))...................-----.....((((...((((((((((((........)))))))))))).-.....))))..... ( -16.30, z-score = -1.93, R) >droGri2.scaffold_15110 14716949 95 - 24565398 GAAUUUUAUUUCGAU-UUCUUUUCGUGCUGCUGCUUGGAGAGACAGUUUGCUUUUGAUUUUCAAAUUGCCUUUUGCUUGGCAAUUCUCUGAAUUUU---------- (((..(((...(((.-.((((((((.((....)).))))))))....)))....)))..))).(((((((........)))))))...........---------- ( -20.30, z-score = -1.14, R) >droSim1.chr3L 21668594 100 - 22553184 GAAUUUCAUUUGAAUGUUUUUCCCUUAUUUCUAUUUGUAUC-----UUGUUACUCGAUUUGGAAAUUGCCUUUUGCCCGGUAAUUUUCAGU-UUUUAGAGUUUUGU (((...(((....)))...)))...................-----.....((((...((((((((((((........)))))))))))).-.....))))..... ( -16.30, z-score = -1.68, R) >droSec1.super_11 2270290 100 - 2888827 GAAUUUCAUUUGAAUGUUUUUCCCUUAUUUCUAUUUGUAUC-----UUGUUACUCGAUUUUGAAAUUGCCUUUUGCCCGGUAAUUUUCAGU-UUUUAGAGUUUUGU (((...(((....)))...)))...................-----.....((((....(((((((((((........))))).)))))).-.....))))..... ( -12.50, z-score = -0.97, R) >droYak2.chr3L 22747123 100 - 24197627 GAAUUUCAUUUGAAUGUUUUUCCCUUAUUUUUAUUUAUAUC-----UUGUUACUCCAUUUGGAAAUUGCCUUUUGCCUGGUAAUUUUCAGU-UUUUAGAGUUUUGU (((...(((....)))...)))...................-----.....((((...((((((((((((........)))))))))))).-.....))))..... ( -15.80, z-score = -2.03, R) >droEre2.scaffold_4784 22004614 100 - 25762168 GAAUUUCAUUUGAAUGUUUUUCUCUUAUUUUUAUUUAUAUC-----UUGUUACUCGAAUUGGAAAUUGCCUUUUGCCUGGUAAUUUUCAGU-UUUUAGAGUUUUGU (((...(((....)))...)))...................-----.....(((((((((((((((((((........)))))))))))))-))...))))..... ( -20.00, z-score = -3.29, R) >droAna3.scaffold_13337 22073403 104 - 23293914 GAAUUUCAUUUGUAUGUUUUCCUCUUAUUUUUAUUUAUAUCG--CGCCGCUUCUCGUUUUGGAAAUUGCCUUUUGCCUGGUAAUUUUCAGUAUUUUAGAGUUUUGA (((...(((....)))..)))((((................(--((........))).((((((((((((........))))))))))))......))))...... ( -16.10, z-score = -1.07, R) >consensus GAAUUUCAUUUGAAUGUUUUUCCCUUAUUUUUAUUUGUAUC_____UUGUUACUCGAUUUGGAAAUUGCCUUUUGCCUGGUAAUUUUCAGU_UUUUAGAGUUUUGU ...................................................((((...((((((((((((........)))))))))))).......))))..... (-10.89 = -12.20 + 1.31)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:44:28 2011