| Sequence ID | dm3.chr3L |
|---|---|
| Location | 22,329,752 – 22,329,864 |
| Length | 112 |
| Max. P | 0.958737 |

| Location | 22,329,752 – 22,329,864 |
|---|---|
| Length | 112 |
| Sequences | 7 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 73.07 |
| Shannon entropy | 0.53156 |
| G+C content | 0.49942 |
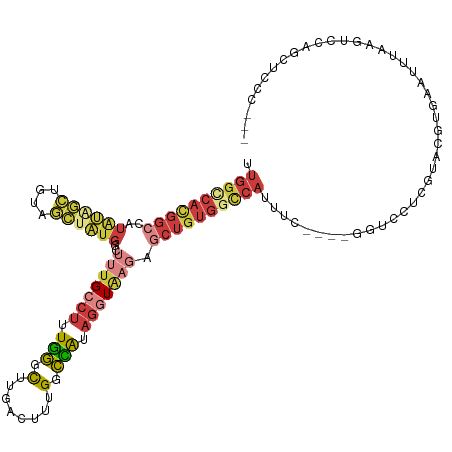
| Mean single sequence MFE | -32.53 |
| Consensus MFE | -11.85 |
| Energy contribution | -14.80 |
| Covariance contribution | 2.95 |
| Combinations/Pair | 1.34 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.36 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.561303 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
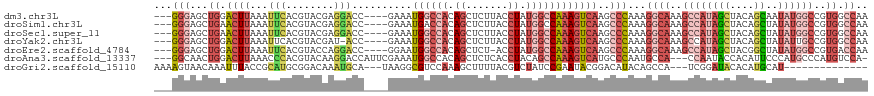
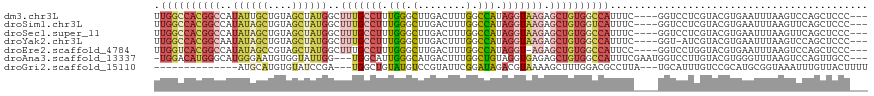
>dm3.chr3L 22329752 112 + 24543557 ---GGGAGCUGGACUUAAAUUCACGUACGAGGACC----GAAAUGGCCACAGCUCUUACCUAUGGCCAAAGUCAAGCCCAAAGGCAAAGCCAUAGCUACAGCAAUAUGGCCGUGGCCAA ---((((((((.........((......))((.((----.....)))).)))))))).....((((((.......(((....)))...((((((((....))..))))))..)))))). ( -37.80, z-score = -2.07, R) >droSim1.chr3L 21664116 112 + 22553184 ---GGGAGCUGAACUUAAAUUCACGUACGAGGACC----GAAAUGACCACAGCUCUUACCUAUGGCCAAAGUCAAGCCCAAAGGCAAAGCCAUAGCUACAGCUAUAUGGCCGUGGCCAA ---((((((((.........((......))((...----.......)).)))))))).....((((((..((((.(((....)))......(((((....))))).))))..)))))). ( -35.40, z-score = -2.41, R) >droSec1.super_11 2265817 112 + 2888827 ---GGGAGCUGAACUUAAAUUCACGUACGAGGACC----GAAAUGGCCACAGCUCUUACCUAUGGCCAAAGUCAAGCCCAAAGGCAAAGCCAUAGCUACAGCUAUAUGGCCGUGGCCAA ---((((((((.........((......))((.((----.....)))).)))))))).....((((((..((((.(((....)))......(((((....))))).))))..)))))). ( -39.20, z-score = -2.79, R) >droYak2.chr3L 22741414 111 + 24197627 ---GGGAGCUGGACUUAAAUUCACGUACGAU-ACC----GAAAUGGCCACAGCUCUUACCUAUGGCCAAAGUCAAGCCCAAAGGCAAAGCCAUAGCUACAGCUAUAUUGCCGUGGCCAA ---.((.(((.(((((...........((..-..)----)...((((((.((.......)).))))))))))).)))))...(((...)))...(((((.((......)).)))))... ( -34.50, z-score = -1.81, R) >droEre2.scaffold_4784 22000252 111 + 25762168 ---GGGAGCUGGACUUAAAUUCACGUACCAGGACC----GGAAUGGCCACAGCUCU-ACCUAUGGCCAAAGUCAAGCCCAAAGGCAAAGCCAUAGCUACGGCUAUAUGGCCGUGACCAA ---.((.(((.(((((...........((......----))..((((((.((....-..)).))))))))))).)))))...((((..((((((((....))))..))))..)).)).. ( -40.60, z-score = -2.60, R) >droAna3.scaffold_13337 22068897 112 + 23293914 ---GGCAACUGGACUUAAACCCACGUACAAGGACCAUUCGAAAUGGCCACAGCUCUCACCUACAGCCAAAGUCAUGCCCAAUGCCA---CCAAUACCACAUUCCCAUGCCCAUGUCCA- ---(((((((((........))).))....((..((((.(..(((((....(((.........)))....)))))..).))))...---))...............))))........- ( -17.10, z-score = 0.76, R) >droGri2.scaffold_15110 14710726 99 + 24565398 AAAAGUAACAAAUUUACCGCAUGCGGACAAAUGCA---UAAGGCGUCCAAAGCUUUUACGUCUAUCCGAAUACGGACAUACAGCCA---UCGGAUACACAUGCAU-------------- ................(((....)))....(((((---((((((.......)))))...((.(((((((....((........)).---))))))).))))))))-------------- ( -23.10, z-score = -2.18, R) >consensus ___GGGAGCUGGACUUAAAUUCACGUACGAGGACC____GAAAUGGCCACAGCUCUUACCUAUGGCCAAAGUCAAGCCCAAAGGCAAAGCCAUAGCUACAGCUAUAUGGCCGUGGCCAA ....((.(((..((((...(((........)))..........((((((.((.......)).))))))))))..)))))...((((..((((((((....))..))))))..)).)).. (-11.85 = -14.80 + 2.95)
| Location | 22,329,752 – 22,329,864 |
|---|---|
| Length | 112 |
| Sequences | 7 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 73.07 |
| Shannon entropy | 0.53156 |
| G+C content | 0.49942 |
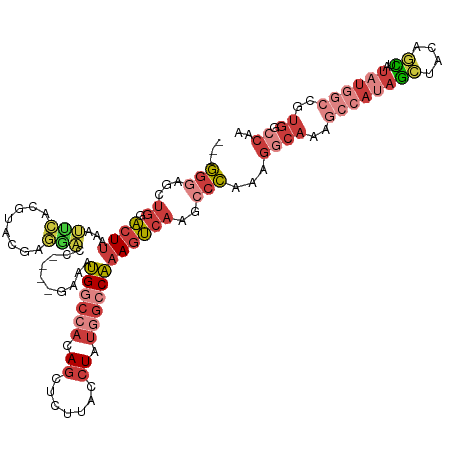
| Mean single sequence MFE | -38.14 |
| Consensus MFE | -22.15 |
| Energy contribution | -24.16 |
| Covariance contribution | 2.01 |
| Combinations/Pair | 1.52 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.66 |
| SVM RNA-class probability | 0.958737 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 22329752 112 - 24543557 UUGGCCACGGCCAUAUUGCUGUAGCUAUGGCUUUGCCUUUGGGCUUGACUUUGGCCAUAGGUAAGAGCUGUGGCCAUUUC----GGUCCUCGUACGUGAAUUUAAGUCCAGCUCCC--- ..(((...(((((((..((....)))))))))..)))...(((((.((((((((((((((.......))))))))).(((----(((......)).))))...))))).)))))..--- ( -40.60, z-score = -1.78, R) >droSim1.chr3L 21664116 112 - 22553184 UUGGCCACGGCCAUAUAGCUGUAGCUAUGGCUUUGCCUUUGGGCUUGACUUUGGCCAUAGGUAAGAGCUGUGGUCAUUUC----GGUCCUCGUACGUGAAUUUAAGUUCAGCUCCC--- .((((((((((..((((((....))))))..(((((((...((((.......))))..))))))).))))))))))....----............(((((....)))))......--- ( -38.50, z-score = -1.59, R) >droSec1.super_11 2265817 112 - 2888827 UUGGCCACGGCCAUAUAGCUGUAGCUAUGGCUUUGCCUUUGGGCUUGACUUUGGCCAUAGGUAAGAGCUGUGGCCAUUUC----GGUCCUCGUACGUGAAUUUAAGUUCAGCUCCC--- .((((((((((..((((((....))))))..(((((((...((((.......))))..))))))).))))))))))....----............(((((....)))))......--- ( -41.20, z-score = -2.02, R) >droYak2.chr3L 22741414 111 - 24197627 UUGGCCACGGCAAUAUAGCUGUAGCUAUGGCUUUGCCUUUGGGCUUGACUUUGGCCAUAGGUAAGAGCUGUGGCCAUUUC----GGU-AUCGUACGUGAAUUUAAGUCCAGCUCCC--- .((((((((((..((((((....))))))..(((((((...((((.......))))..))))))).)))))))))).(((----(((-(...))).))))................--- ( -40.40, z-score = -1.92, R) >droEre2.scaffold_4784 22000252 111 - 25762168 UUGGUCACGGCCAUAUAGCCGUAGCUAUGGCUUUGCCUUUGGGCUUGACUUUGGCCAUAGGU-AGAGCUGUGGCCAUUCC----GGUCCUGGUACGUGAAUUUAAGUCCAGCUCCC--- ..((..(((((......))))).(((((((((((((((...((((.......))))..))))-)))))))))))....))----((..((((.((..........))))))..)).--- ( -46.20, z-score = -2.99, R) >droAna3.scaffold_13337 22068897 112 - 23293914 -UGGACAUGGGCAUGGGAAUGUGGUAUUGG---UGGCAUUGGGCAUGACUUUGGCUGUAGGUGAGAGCUGUGGCCAUUUCGAAUGGUCCUUGUACGUGGGUUUAAGUCCAGUUGCC--- -(((((.(((((.....(((((.(......---).)))))((((.(((...((((..(((.(....))))..))))..)))....))))..........))))).)))))......--- ( -31.20, z-score = 0.74, R) >droGri2.scaffold_15110 14710726 99 - 24565398 --------------AUGCAUGUGUAUCCGA---UGGCUGUAUGUCCGUAUUCGGAUAGACGUAAAAGCUUUGGACGCCUUA---UGCAUUUGUCCGCAUGCGGUAAAUUUGUUACUUUU --------------.((((((.(..(((((---.((((((.((((((....)))))).)).....)))))))))..)..))---))))((((.(((....)))))))............ ( -28.90, z-score = -1.78, R) >consensus UUGGCCACGGCCAUAUAGCUGUAGCUAUGGCUUUGCCUUUGGGCUUGACUUUGGCCAUAGGUAAGAGCUGUGGCCAUUUC____GGUCCUCGUACGUGAAUUUAAGUCCAGCUCCC___ .((((((((((..((((((....))))))..(((((((.(((.(........).))).))))))).))))))))))........................................... (-22.15 = -24.16 + 2.01)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:44:26 2011