| Sequence ID | dm3.chr2L |
|---|---|
| Location | 6,133,975 – 6,134,066 |
| Length | 91 |
| Max. P | 0.559023 |

| Location | 6,133,975 – 6,134,066 |
|---|---|
| Length | 91 |
| Sequences | 10 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 69.88 |
| Shannon entropy | 0.63063 |
| G+C content | 0.37391 |
| Mean single sequence MFE | -18.33 |
| Consensus MFE | -7.08 |
| Energy contribution | -8.01 |
| Covariance contribution | 0.93 |
| Combinations/Pair | 1.52 |
| Mean z-score | -1.26 |
| Structure conservation index | 0.39 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.559023 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
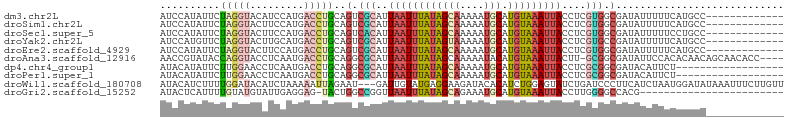
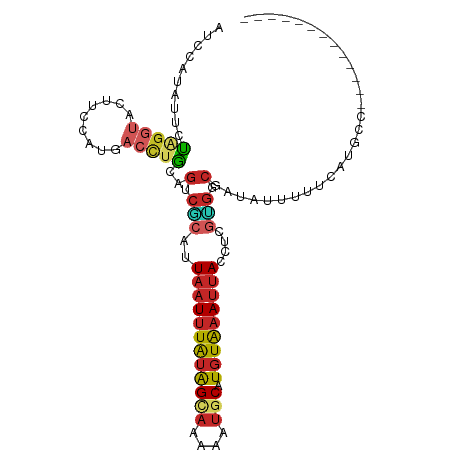
>dm3.chr2L 6133975 91 + 23011544 AUCCAUAUUCUAGGUACAUCCAUGACCUGCAGUCGCAUUAAUUUAUAGCAAAAAUGCAUGUAAAUUACCUCGUGGCGAUAUUUUUCAUGCC------------- ...........((((.((....))))))((((((((..((((((((((((....))).)))))))))....)))))((......)).))).------------- ( -19.20, z-score = -1.73, R) >droSim1.chr2L 5921472 91 + 22036055 AUCCAUAUUCUAGGUACUUCCAUGACCUGCAGUCGCAUUAAUUUAUAGCAAAAAUGCAUGUAAAUUACCUCGUGGCGAUAUUUUUCAUGCC------------- ...........((((.(......)))))((((((((..((((((((((((....))).)))))))))....)))))((......)).))).------------- ( -19.00, z-score = -1.78, R) >droSec1.super_5 4204189 91 + 5866729 AUCCAUAUUCUAGGUACUUCCAUGACCUGCAGUCACAUUAAUUUAUAGCAAAAAUGCAUGUAAAUUACCUCGUGGCGAUAUUUUUCCUGCC------------- ...........((((.(......)))))((((((((..((((((((((((....))).)))))))))....)))))((......)).))).------------- ( -18.30, z-score = -2.03, R) >droYak2.chr2L 15544526 91 - 22324452 AUCCAUGUUCUAGGUACUUGCAUGACCUGCAGUCGCAUUAAUUUAUAGUAAAAAUGCAUGUAAAUUACCUCGUGCCGAUAUUUUUCAUGCC------------- ...((((....(((((...((((((..(((....))).((((((((((((....))).)))))))))..))))))...)))))..))))..------------- ( -18.10, z-score = -1.26, R) >droEre2.scaffold_4929 15057938 91 + 26641161 AUCCAUAUUCUAGGUACUUCCAUGACCUGCAGUCGCAUUAAUUUAUAGCAAAAAUGCAUGUAAAUUACCUCGUGGCGAUAUUUUUCAUGCC------------- ...........((((.(......)))))((((((((..((((((((((((....))).)))))))))....)))))((......)).))).------------- ( -19.00, z-score = -1.78, R) >droAna3.scaffold_12916 8120367 99 - 16180835 AACCGUAUACCAGGUACCUCAAUGACCUGCAGGCGCAUUAAUUUAUAGCAAAAAUACAUGUAAAUUACUU-GCGGCGAUAUUCCACACAACAGCAACACC---- ....((....(((((.........)))))..(.((((.(((((((((...........)))))))))..)-))).)................))......---- ( -17.40, z-score = -0.71, R) >dp4.chr4_group1 52396 86 + 5278887 AUACAUAUUCUUGGAACCUCAAUGACCUGCAGGCGCAUUAAUUUAUAGCAAAAAUGCAUGUAAAUUACCUCGCGGCGAUACAUUCU------------------ ....................((((...(((..(((...((((((((((((....))).)))))))))...))).)))...))))..------------------ ( -15.70, z-score = -0.63, R) >droPer1.super_1 51465 86 + 10282868 AUACAUAUUCUUGGAACCUCAAUGACCUGCAGGCGCAUUAAUUUAUAGCAAAAAUGCAUGUAAAUUACCUCGCGGCGAUACAUUCU------------------ ....................((((...(((..(((...((((((((((((....))).)))))))))...))).)))...))))..------------------ ( -15.70, z-score = -0.63, R) >droWil1.scaffold_180708 7659293 101 + 12563649 AUACAUCUUUUGGAUACAUCUAAAAAUUAGAAU---GAUUGUAUGAGGAAGAUACACAUCUGGAGUAUCUGAUCCCUUCAUCUAAUGGAUAUAAAUUUCUUGUU ....((((.((((((.((((((.....))).))---).......((((.((((((.(.....).))))))....)))).)))))).)))).............. ( -17.40, z-score = 0.49, R) >droGri2.scaffold_15252 2752296 79 + 17193109 AUACUCAUUUUGUAUGUAUUGAGGAG-UACUGGCCGGUUAAUUUAUAGCAGAAAUGCAUGUAAAUUACCUUGGGGCCACG------------------------ .(((((.(((.((....)).))))))-)).((((((((.(((((((((((....))).)))))))))))....)))))..------------------------ ( -23.50, z-score = -2.54, R) >consensus AUCCAUAUUCUAGGUACUUCCAUGACCUGCAGUCGCAUUAAUUUAUAGCAAAAAUGCAUGUAAAUUACCUCGUGGCGAUAUUUUUCAUGCC_____________ ..........(((((.........)))))..(.(((..((((((((((((....))).)))))))))....))).)............................ ( -7.08 = -8.01 + 0.93)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:20:27 2011