| Sequence ID | dm3.chr3L |
|---|---|
| Location | 22,305,678 – 22,305,812 |
| Length | 134 |
| Max. P | 0.868161 |

| Location | 22,305,678 – 22,305,812 |
|---|---|
| Length | 134 |
| Sequences | 5 |
| Columns | 134 |
| Reading direction | forward |
| Mean pairwise identity | 86.12 |
| Shannon entropy | 0.24377 |
| G+C content | 0.59676 |
| Mean single sequence MFE | -57.02 |
| Consensus MFE | -42.98 |
| Energy contribution | -43.46 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.75 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.99 |
| SVM RNA-class probability | 0.868161 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
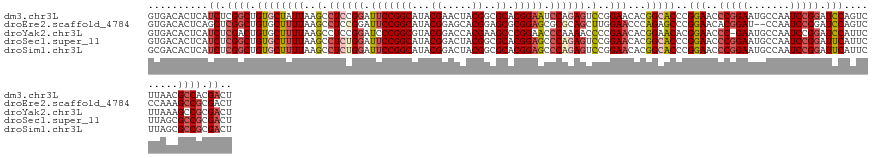
>dm3.chr3L 22305678 134 + 24543557 AGUCGUGGCGUUAAGACUGGAUCCGGAUUGGCAUUCCGGGUUCCGGGUGCCGUGUUCCGGACUCUGGAUUCCGUGCGCCGUAGUUCGUAUGCCGGAAUCCGGAGGCUUAAUAGCACAGCCGAGAUGAGUGUCAC .(((.((((............((((((.((((((.((((...))))))))))...))))))((((((((((((.(((.((.....))..)))))))))))))))(((....)))...)))).)))......... ( -59.50, z-score = -3.03, R) >droEre2.scaffold_4784 21976542 132 + 25762168 AGUCGCGGCUUUGGGACUGGAUCCGGAUUGG--AUCCGUGUUCCGGGCUCUGGGUUCCAAGCUGCGCGCUCCGCGCUCCGUGCUCCGUAUGCCGGAAUCCGGAGGCUUAAAAGCACAGCCGAGCUGAGUGUCAC .(.(((.(((((((((((((((((.....))--))))).)))))((.(((((((((((..(..(((((...)))))..)(.((.......)))))))))))))).))..))))).((((...)))).))).).. ( -60.70, z-score = -1.92, R) >droYak2.chr3L 22706848 133 + 24197627 AGUCGCGGCUUUAAGAAUGGAUCCGGAUUGGCAUUC-GGGUUCCGUGUUCCGUGUUCGGGGUUUUGGGUUCCGGGCUUCGUGGUCCGUACGCCGGGAUCCGGAGGCUUAAAAGCACAGUCGAGAUGAGUGUCAC .(((.(((((......((((((((((((....))))-))).))))).....(((((..(((((((.((.(((.(((..((.....))...))).))).)).)))))))...)))))))))).)))......... ( -48.00, z-score = -0.48, R) >droSec1.super_11 2241310 134 + 2888827 AGUCGCGGCGCUAAGAAUGAAUCCGGAUUGGCAUUCCGGGUUCCGGGUGCCGUGUUCCGGACUCUGGGCUCCGUGCGCCGUAGUCCGUAUGCCGGAAUCCAGAGGCUUAAAAGCACAGCCGAGAUGAGUGUCAC .(((.((((............((((((.((((((.((((...))))))))))...))))))((((((..((((.(((.((.....))..)))))))..))))))(((....)))...)))).)))......... ( -57.80, z-score = -2.45, R) >droSim1.chr3L 21635994 134 + 22553184 AGUCGCGGCGCUAAGAAUGAAUCCGGAUUGGCAUUCCGGGUUCCGGGUGCCGUGUUCCGGACUCUGGGCUCCGUGCGCCGUAGUCCGUAUGCCGGAAUCCAGAGGCUUAAAAGCACAGCCGAGAUGAGUGUCGC ....((((((((.........((((((.((((((.((((...))))))))))...))))))((((((..((((.(((.((.....))..)))))))..))))))(((....)))............)))))))) ( -59.10, z-score = -2.47, R) >consensus AGUCGCGGCGUUAAGAAUGGAUCCGGAUUGGCAUUCCGGGUUCCGGGUGCCGUGUUCCGGACUCUGGGCUCCGUGCGCCGUAGUCCGUAUGCCGGAAUCCGGAGGCUUAAAAGCACAGCCGAGAUGAGUGUCAC .(((.((((............((((((.(((((.((((.....)))))))))...))))))(((((((.((((.((..((.....))...)))))).)))))))(((....)))...)))).)))......... (-42.98 = -43.46 + 0.48)


| Location | 22,305,678 – 22,305,812 |
|---|---|
| Length | 134 |
| Sequences | 5 |
| Columns | 134 |
| Reading direction | reverse |
| Mean pairwise identity | 86.12 |
| Shannon entropy | 0.24377 |
| G+C content | 0.59676 |
| Mean single sequence MFE | -47.80 |
| Consensus MFE | -31.10 |
| Energy contribution | -32.98 |
| Covariance contribution | 1.88 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.549350 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 22305678 134 - 24543557 GUGACACUCAUCUCGGCUGUGCUAUUAAGCCUCCGGAUUCCGGCAUACGAACUACGGCGCACGGAAUCCAGAGUCCGGAACACGGCACCCGGAACCCGGAAUGCCAAUCCGGAUCCAGUCUUAACGCCACGACU ..(((.(((.....((((.........))))...((((((((((...((.....))..)).)))))))).)))((((((....((((.((((...))))..))))..))))))....))).............. ( -48.10, z-score = -2.60, R) >droEre2.scaffold_4784 21976542 132 - 25762168 GUGACACUCAGCUCGGCUGUGCUUUUAAGCCUCCGGAUUCCGGCAUACGGAGCACGGAGCGCGGAGCGCGCAGCUUGGAACCCAGAGCCCGGAACACGGAU--CCAAUCCGGAUCCAGUCCCAAAGCCGCGACU (((...((..((((((((((((.(((..(((((((..(((((.....)))))..))))).)).))).))))))))(((...)))))))..))..)))((((--((.....))))))((((.(......).)))) ( -55.70, z-score = -2.42, R) >droYak2.chr3L 22706848 133 - 24197627 GUGACACUCAUCUCGACUGUGCUUUUAAGCCUCCGGAUCCCGGCGUACGGACCACGAAGCCCGGAACCCAAAACCCCGAACACGGAACACGGAACCC-GAAUGCCAAUCCGGAUCCAUUCUUAAAGCCGCGACU ((((((.((.....)).)))(((((.(((..(((((((...(((((.(((.((.((.....))............(((....))).....))...))-).))))).)))))))......))))))))))).... ( -33.80, z-score = -0.51, R) >droSec1.super_11 2241310 134 - 2888827 GUGACACUCAUCUCGGCUGUGCUUUUAAGCCUCUGGAUUCCGGCAUACGGACUACGGCGCACGGAGCCCAGAGUCCGGAACACGGCACCCGGAACCCGGAAUGCCAAUCCGGAUUCAUUCUUAGCGCCGCGACU ..........((.((((.(((((....)))((((((..((((((...((.....))..)).))))..))))))((((((....((((.((((...))))..))))..))))))..........)))))).)).. ( -50.30, z-score = -2.12, R) >droSim1.chr3L 21635994 134 - 22553184 GCGACACUCAUCUCGGCUGUGCUUUUAAGCCUCUGGAUUCCGGCAUACGGACUACGGCGCACGGAGCCCAGAGUCCGGAACACGGCACCCGGAACCCGGAAUGCCAAUCCGGAUUCAUUCUUAGCGCCGCGACU .(((........)))((.(((((.......((((((..((((((...((.....))..)).))))..))))))((((((....((((.((((...))))..))))..)))))).........))))).)).... ( -51.10, z-score = -2.37, R) >consensus GUGACACUCAUCUCGGCUGUGCUUUUAAGCCUCCGGAUUCCGGCAUACGGACUACGGCGCACGGAGCCCAGAGUCCGGAACACGGCACCCGGAACCCGGAAUGCCAAUCCGGAUCCAUUCUUAACGCCGCGACU ..........((.((((.((((((((..(.((((((.(((((((...((.....))..)).))))).)))))).)..)))...)))))..(((..(((((.......))))).))).........)))).)).. (-31.10 = -32.98 + 1.88)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:44:18 2011