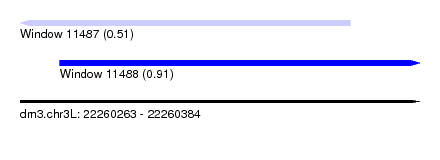
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 22,260,263 – 22,260,384 |
| Length | 121 |
| Max. P | 0.907302 |

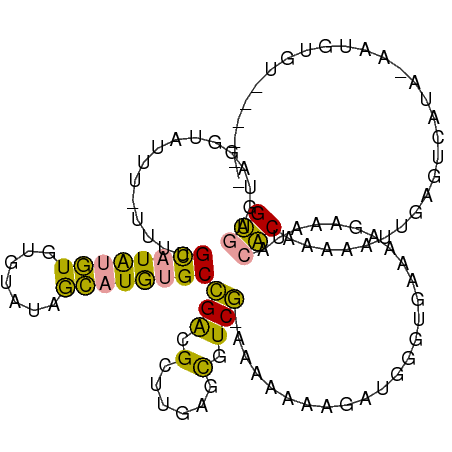
| Location | 22,260,263 – 22,260,363 |
|---|---|
| Length | 100 |
| Sequences | 3 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 57.48 |
| Shannon entropy | 0.58288 |
| G+C content | 0.42303 |
| Mean single sequence MFE | -28.37 |
| Consensus MFE | -9.25 |
| Energy contribution | -7.60 |
| Covariance contribution | -1.65 |
| Combinations/Pair | 1.60 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.33 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.508733 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 22260263 100 - 24543557 GGCCGCAGGUCAUU-UUCUGUAUGCAUAAAAUUUGUAUGGGCGGAAGCGAGAUUGUCU-AAGAAAUGAAGGGUGAAAGGGAACACCCAAGGAAAAGGUCGUG-------------- ((((...((.((.(-(((..((((((.......)))))).((....)))))).)).))-..........(((((........)))))........))))...-------------- ( -28.10, z-score = -2.19, R) >droWil1.scaffold_181153 18170 115 - 180639 GUGAGUAGGUAUUU-UUUCGCAUAUGUGUGUACAGCAUGUGCCGGCGAUUGAGCCUCGAAAAAAAAGAUGGGUGAAAAGACAUACACAAAAAUUGAGUCAUAGAAUGUGUAGAGUG ((((.(..((.(((-((((((((((((.......)))))))).(((......)))..)))))))...(((..(....)..)))........))..).))))............... ( -24.30, z-score = -0.49, R) >droMoj3.scaffold_3993 7928 110 + 10554 GUGAGUAGGUAUUUCUUUUGCAUAUGUGUGUGUGACAUAUGCCGACGCUUGAGCGUCG-CAAGAAAGUUGGGUGAAAAGAGAUACACAAAAAUUGAAGCAUACAAUGUGUG----- ........(((((((((((((((((((.......))))))))...((((..(...((.-...))...)..)))))))))))))))(((...((((.......)))).))).----- ( -32.70, z-score = -2.60, R) >consensus GUGAGUAGGUAUUU_UUUUGCAUAUGUGUGUAUAGCAUGUGCCGACGCUUGAGCGUCG_AAAAAAAGAUGGGUGAAAAGAAAUACACAAAAAUUGAGUCAUA_AAUGUGU______ (((................((((((((.......))))))))(((.(......).)))..........................)))............................. ( -9.25 = -7.60 + -1.65)

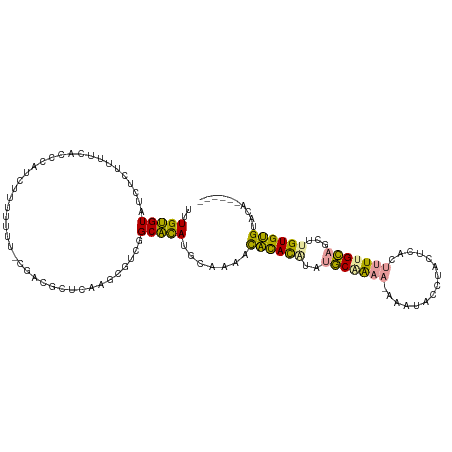
| Location | 22,260,275 – 22,260,384 |
|---|---|
| Length | 109 |
| Sequences | 3 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 61.29 |
| Shannon entropy | 0.52963 |
| G+C content | 0.43593 |
| Mean single sequence MFE | -26.75 |
| Consensus MFE | -10.32 |
| Energy contribution | -9.34 |
| Covariance contribution | -0.98 |
| Combinations/Pair | 1.56 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.39 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.19 |
| SVM RNA-class probability | 0.907302 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 22260275 109 + 24543557 CUUGGGUGUUCCCUUUCACCCUUCAUUUCUU-AGACAAUCUCGCUUCCGCCCAUACAAAUUUUAUGCAUACAGAA-AAUGACCUGCGGCCGAUCGUUGCUUGUGAGUCGCU----- ...(((((........)))))(((((.....-((.((((.(((...((((........(((((.((....)).))-))).....)))).)))..)))))).))))).....----- ( -20.42, z-score = 0.13, R) >droWil1.scaffold_181153 18196 115 + 180639 UUUGUGUAUGUCUUUUCACCCAUCUUUUUUUUCGAGGCUCAAUCGCCGGCACAUGCUGUACACACAUAUGCGAAA-AAAUACCUACUCACUUUUGCAGCUUGUGUGUACAAACACU ...((((.((((..........((.........))(((......))))))).....((((((((((..(((((((-..............)))))))...)))))))))).)))). ( -30.14, z-score = -3.36, R) >droMoj3.scaffold_3993 7949 109 - 10554 UUUGUGUAUCUCUUUUCACCCAACUUUCUUG-CGACGCUCAAGCGUCGGCAUAUGUCACACACACAUAUGCAAAAGAAAUACCUACUCACUUUUGCAGCUAGUGUGCACA------ ..((((((................((((((.-((((((....))))))((((((((.......))))))))..))))))........((((.........))))))))))------ ( -29.70, z-score = -3.50, R) >consensus UUUGUGUAUCUCUUUUCACCCAUCUUUUUUU_CGACGCUCAAGCGUCGGCACAUGCAAAACACACAUAUGCAAAA_AAAUACCUACUCACUUUUGCAGCUUGUGUGUACA______ ..(((((.........................................))))).......((((((..(((((((...............)))))))...)))))).......... (-10.32 = -9.34 + -0.98)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:44:14 2011