
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 22,226,303 – 22,226,398 |
| Length | 95 |
| Max. P | 0.646835 |

| Location | 22,226,303 – 22,226,398 |
|---|---|
| Length | 95 |
| Sequences | 9 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 77.20 |
| Shannon entropy | 0.46107 |
| G+C content | 0.51449 |
| Mean single sequence MFE | -29.94 |
| Consensus MFE | -16.38 |
| Energy contribution | -16.18 |
| Covariance contribution | -0.21 |
| Combinations/Pair | 1.42 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.646835 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 22226303 95 + 24543557 ---UUAUUAUUGUUG-UUGGUCAGUUAUUAUGGCCACUGCAAUUUUGUAGCGCUUCAUGUCCGG---CUACGGGCAGGAGUCCCUCCUCCGGUCCUGAAUUC ---.....(((((.(-(.(((((.......))))))).)))))...(((((((.....))...)---))))((((.((((......)))).))))....... ( -28.10, z-score = -1.63, R) >droSim1.chr3L_random 945023 95 + 1049610 ---UUAUUAUUGUUG-UUGGUCAGUUAUUAUGGCCACUGCAAUUUUGUAGCGCUUCAUGUCCGG---CUACGGGCAGGAGUCCCUCCUCCGGCCCUGAAUUC ---.....(((((.(-(.(((((.......))))))).)))))...(((((((.....))...)---))))((((.((((......)))).))))....... ( -30.80, z-score = -2.16, R) >droSec1.super_11 2156892 95 + 2888827 ---UUAUUAUUGUUG-UUGGUCAGUUAUUAUGGCCACUGCAAUUUUGUAGCGCUUCAUGUCCGG---CUACGGGCAGGAGUCCCUCCUCCGGCCCUGAAUUU ---.....(((((.(-(.(((((.......))))))).)))))...(((((((.....))...)---))))((((.((((......)))).))))....... ( -30.80, z-score = -2.09, R) >droYak2.chr3L 22612966 95 + 24197627 ---UUAUUAUUGCUG-UUGGGCAGUUAUUAUGGCCACUGCAAUUUUGUAGCGCUUCAUGUCCGG---CUACGGGCAGGAGCUCCUCCUCCGGCCCUGAAUUC ---.....((((((.-...))))))..(((.((((.(((((....))))).(((((.((((((.---...))))))))))).........)))).))).... ( -33.30, z-score = -1.81, R) >droEre2.scaffold_4784 21896972 95 + 25762168 ---UUAUUAUUGCUG-UUGGUCAGUUAUUAUGGCCACUGCAAUUUUGUAGCGCUUCAUGUCCGG---CGACGGGCAGGAGCUCCUCCACCGGCCCGGAAUUC ---((((.(((((.(-(.(((((.......))))))).)))))...)))).(((((.((((((.---...)))))))))))...(((........))).... ( -33.00, z-score = -1.71, R) >droAna3.scaffold_13337 21387720 94 + 23293914 ---UUGGCAUUGUUG-CCAGUCAGUUAUUACGGGCUCUGCAAUUUUGUAGCGCUUAAUGUCCGGAGUCCGCACUCCGCAGUCCAGCGCCCCUCCCCCA---- ---((((((....))-))))...........((((.(((((....))))).(((..((...((((((....))))))..))..)))))))........---- ( -29.60, z-score = -2.12, R) >dp4.chrXR_group8 6310976 93 + 9212921 ---CUAUUGUUGUUG-UUGGCCAGUUAUUAUGGCCACUGCAAUUUUGUAGCGCUUAAUGUCCGG---CUACGGGUCUGAGCCCCGGCUACGGUUACGUCU-- ---.....(((((.(-(.(((((.......))))))).))))).((((((((((((...((((.---...))))..)))))....)))))))........-- ( -29.10, z-score = -0.87, R) >droPer1.super_45 446934 93 - 618639 ---CUAUUGUUGUUG-UUGGCCAGUUAUUAUGGCCACUGCAAUUUUGUAGCGCUUAAUGUCCGG---CUACGGGUCUGAGCCCCGGCUACGGUUACGUCU-- ---.....(((((.(-(.(((((.......))))))).))))).((((((((((((...((((.---...))))..)))))....)))))))........-- ( -29.10, z-score = -0.87, R) >droWil1.scaffold_180698 153213 89 + 11422946 UUGCUGCUGCCGGUGGCUGGUCAGUUAUUAUGGCUACUGCAAUUUUGUAGCGCUUAAUGUCCGGG--UCCCGGGUUCGGGUUCGAGGUUCG----------- ..(((((.....(..(.((((((.......)))))))..)......)))))..........((((--.((((....)))))))).......----------- ( -25.70, z-score = 0.61, R) >consensus ___UUAUUAUUGUUG_UUGGUCAGUUAUUAUGGCCACUGCAAUUUUGUAGCGCUUCAUGUCCGG___CUACGGGCAGGAGUCCCUCCUCCGGCCCUGAAUU_ ...((((.(((((....((((((.......))))))..)))))...)))).((((...(((((.......)))))..))))..................... (-16.38 = -16.18 + -0.21)

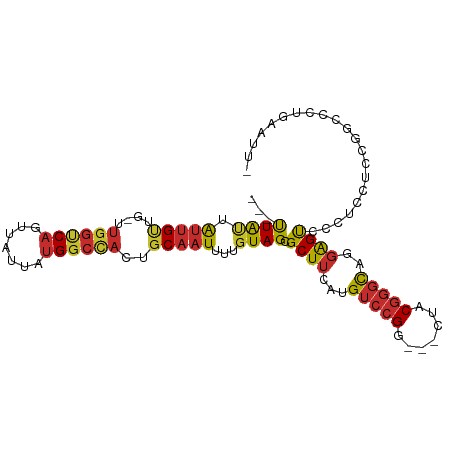

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:44:11 2011