| Sequence ID | dm3.chr3L |
|---|---|
| Location | 22,224,924 – 22,225,026 |
| Length | 102 |
| Max. P | 0.995351 |

| Location | 22,224,924 – 22,225,016 |
|---|---|
| Length | 92 |
| Sequences | 12 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 86.05 |
| Shannon entropy | 0.29558 |
| G+C content | 0.36219 |
| Mean single sequence MFE | -20.58 |
| Consensus MFE | -18.34 |
| Energy contribution | -18.59 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.66 |
| Structure conservation index | 0.89 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.79 |
| SVM RNA-class probability | 0.995351 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
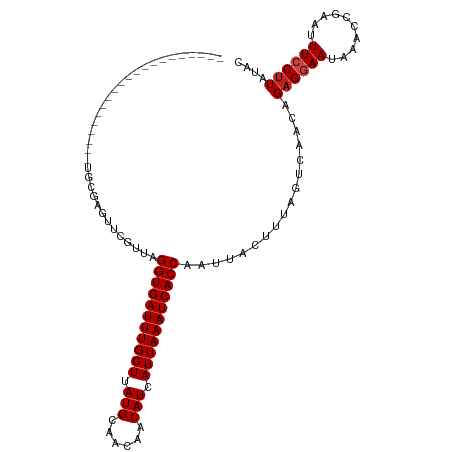
>dm3.chr3L 22224924 92 - 24543557 AGGUGAUUUGGUUAUGCAACAACAUCAUUAAAUCACCAAUUACUUUAGUCAACAGAGGAAUAAACCGAAUUUCCUUAUACACAAGCCGGGUC--------------- .(((((((((((.(((......))).))))))))))).................((((((..........))))))................--------------- ( -20.00, z-score = -2.34, R) >droPer1.super_45 445058 107 + 618639 AGGUGAUUUGGUUAUGCAACAACAUCAUUAAAUCACCAAUUACUUUAGUCAACAGAGGAAUAAACCAAAUUUCCUUAUACACAAGCCAAACGAACAGAGGCAAGGUC .(((((((((((.(((......))).)))))))))))....(((((........((((((..........))))))........(((...........)))))))). ( -25.00, z-score = -3.82, R) >dp4.chrXR_group8 6309092 107 - 9212921 AGGUGAUUUGGUUAUGCAACAACAUCAUUAAAUCACCAAUUACUUUAGUCAACAGAGGAAUAAACCAAAUUUCCUUAUACACAAGCCAAACGAACAGAGGCAAGGUC .(((((((((((.(((......))).)))))))))))....(((((........((((((..........))))))........(((...........)))))))). ( -25.00, z-score = -3.82, R) >droAna3.scaffold_13337 21386224 105 - 23293914 AGGUGAUUUGGUUAUGCAACAACAUCAUUAAAUCACCAAUUACUUUAGUCAACAGAGGAACAAACCGAAUUUCCUUACACACAAGUCAAGUCGCAGCCGGUGCCG-- .(((((((((((.(((......))).)))))))))))...................((.....((((......(((((......)).))).......)))).)).-- ( -21.02, z-score = -1.30, R) >droEre2.scaffold_4784 21895598 92 - 25762168 AGGUGAUUUGGUUAUGCAACAACAUCAUUAAAUCACCAAUUACUUUAGUCAACAGAGGAAUAAACCGAAUUUCCUUCUACACAAGCCGGGUG--------------- .(((((((((((.(((......))).))))))))))).................((((((..........))))))...(((.......)))--------------- ( -19.70, z-score = -1.89, R) >droYak2.chr3L 22611565 92 - 24197627 AGGUGAUUUGGUUAUGCAACAACAUCAUUAAAUCACCAAUUACUUUAGUCAACAGAGGAAUAAACCGAAUUUCCUUAUACACAAGCCGGGUC--------------- .(((((((((((.(((......))).))))))))))).................((((((..........))))))................--------------- ( -20.00, z-score = -2.34, R) >droSec1.super_11 2155492 92 - 2888827 AGGUGAUUUGGUUAUGCAACAACAUCAUUAAAUCACCAAUUACUUUAGUCAACAGAGGAAUAAACCGAAUUUCCUUAUACACAAGCCGGGUC--------------- .(((((((((((.(((......))).))))))))))).................((((((..........))))))................--------------- ( -20.00, z-score = -2.34, R) >droSim1.chr3L_random 943614 92 - 1049610 AGGUGAUUUGGUUAUGCAACAACAUCAUUAAAUCACCAAUUACUUUAGUCAACAGAGGAAUAAACCGAAUUUCCUUAUACACAAGCCGGGUC--------------- .(((((((((((.(((......))).))))))))))).................((((((..........))))))................--------------- ( -20.00, z-score = -2.34, R) >droVir3.scaffold_13049 14791841 89 - 25233164 AGGUGAUUUGGUUAUGCAACAACAUCAUUAAAUCACCAAUUACUUUAGUCAACAGUGGAAUAAAGUGAAUUUCCUUUUACACAAGAGCG------------------ .(((((((((((.(((......))).)))))))))))..((((((((.((.......)).)))))))).....(((((....)))))..------------------ ( -21.70, z-score = -3.21, R) >droMoj3.scaffold_6680 19573797 89 - 24764193 AGGUGAUUUGGUUAUGCAGCAACAUCAUUAAAUCACCAAUUACUUUAGUCAACAGAGAAAUAAACUGAAUUUCCUUAUUCACGAGAGCG------------------ .(((((((((((.(((......))).))))))))))).....((((.((....((.(((((.......))))))).....)).))))..------------------ ( -18.10, z-score = -2.21, R) >droGri2.scaffold_15110 6634524 89 + 24565398 AGGUGAUUUGGUUAUGCAACAACAUCAUUAAAUCACCAAUUACCUUAGUCAACAGAGGAAUAAACUGAAUUUCCUUAUACACAAUGCCC------------------ .(((((((((((.(((......))).))))))))))).................((((((..........)))))).............------------------ ( -20.00, z-score = -2.89, R) >droWil1.scaffold_180698 150946 85 - 11422946 AGGUGAUUUGGUUAUGCAACAACAUCAUUAAAUCACCAAUUACUUUAGUCAACAGAGAAAUAAACCAAAUUUCCUUAUACACAAG---------------------- .(((((((((((.(((......))).)))))))))))..........((....((.(((((.......)))))))...)).....---------------------- ( -16.40, z-score = -3.46, R) >consensus AGGUGAUUUGGUUAUGCAACAACAUCAUUAAAUCACCAAUUACUUUAGUCAACAGAGGAAUAAACCGAAUUUCCUUAUACACAAGCCGGGUC_______________ .(((((((((((.(((......))).))))))))))).................((((((..........))))))............................... (-18.34 = -18.59 + 0.25)


| Location | 22,224,936 – 22,225,026 |
|---|---|
| Length | 90 |
| Sequences | 12 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 85.77 |
| Shannon entropy | 0.29195 |
| G+C content | 0.35866 |
| Mean single sequence MFE | -23.26 |
| Consensus MFE | -18.34 |
| Energy contribution | -18.59 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.53 |
| Structure conservation index | 0.79 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.27 |
| SVM RNA-class probability | 0.987133 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 22224936 90 - 24543557 -----------------------UGCGUUCGUUAGGUGAUUUGGUUAUGCAACAACAUCAUUAAAUCACCAAUUACUUUAGUCAACAGAGGAAUAAACCGAAUUUCCUUAUAC -----------------------...........(((((((((((.(((......))).))))))))))).................((((((..........)))))).... ( -20.00, z-score = -2.66, R) >droPer1.super_45 445085 108 + 618639 ----UCUGUGGCAGUGCC-ACUGUGAGUUCGUUAGGUGAUUUGGUUAUGCAACAACAUCAUUAAAUCACCAAUUACUUUAGUCAACAGAGGAAUAAACCAAAUUUCCUUAUAC ----((((((((...)))-((((.((((..(((.(((((((((((.(((......))).)))))))))))))).))))))))..)))))((((..........))))...... ( -28.20, z-score = -2.59, R) >dp4.chrXR_group8 6309119 108 - 9212921 ----UCUGUGGCAGUGCC-ACUGUGAGUUCGUUAGGUGAUUUGGUUAUGCAACAACAUCAUUAAAUCACCAAUUACUUUAGUCAACAGAGGAAUAAACCAAAUUUCCUUAUAC ----((((((((...)))-((((.((((..(((.(((((((((((.(((......))).)))))))))))))).))))))))..)))))((((..........))))...... ( -28.20, z-score = -2.59, R) >droAna3.scaffold_13337 21386249 105 - 23293914 -----CUUUGGC--UGCC-ACUGCCAGUUCGUUAGGUGAUUUGGUUAUGCAACAACAUCAUUAAAUCACCAAUUACUUUAGUCAACAGAGGAACAAACCGAAUUUCCUUACAC -----...((((--....-...))))((((....(((((((((((.(((......))).))))))))))).....((((.(....)))))))))................... ( -25.20, z-score = -2.42, R) >droEre2.scaffold_4784 21895610 92 - 25762168 ---------------------UGCGAGUUCGUUAGGUGAUUUGGUUAUGCAACAACAUCAUUAAAUCACCAAUUACUUUAGUCAACAGAGGAAUAAACCGAAUUUCCUUCUAC ---------------------.(.(((((((...(((((((((((.(((......))).))))))))))).....((((.(....).)))).......))))))).)...... ( -21.10, z-score = -2.61, R) >droYak2.chr3L 22611577 92 - 24197627 ---------------------UGCGAGUUCGUUAGGUGAUUUGGUUAUGCAACAACAUCAUUAAAUCACCAAUUACUUUAGUCAACAGAGGAAUAAACCGAAUUUCCUUAUAC ---------------------.(((....)))..(((((((((((.(((......))).))))))))))).................((((((..........)))))).... ( -21.20, z-score = -2.82, R) >droSec1.super_11 2155504 92 - 2888827 ---------------------UGCGAGUUCGUUAGGUGAUUUGGUUAUGCAACAACAUCAUUAAAUCACCAAUUACUUUAGUCAACAGAGGAAUAAACCGAAUUUCCUUAUAC ---------------------.(((....)))..(((((((((((.(((......))).))))))))))).................((((((..........)))))).... ( -21.20, z-score = -2.82, R) >droSim1.chr3L_random 943626 92 - 1049610 ---------------------UGCGAGUUCGUUAGGUGAUUUGGUUAUGCAACAACAUCAUUAAAUCACCAAUUACUUUAGUCAACAGAGGAAUAAACCGAAUUUCCUUAUAC ---------------------.(((....)))..(((((((((((.(((......))).))))))))))).................((((((..........)))))).... ( -21.20, z-score = -2.82, R) >droVir3.scaffold_13049 14791850 94 - 25233164 -------------------GUUGCGACUUCGUUAGGUGAUUUGGUUAUGCAACAACAUCAUUAAAUCACCAAUUACUUUAGUCAACAGUGGAAUAAAGUGAAUUUCCUUUUAC -------------------...(((....)))..(((((((((((.(((......))).)))))))))))..((((((((.((.......)).))))))))............ ( -22.60, z-score = -2.60, R) >droMoj3.scaffold_6680 19573806 113 - 24764193 UAGCUGACUGUCAACUGCGACUGCGACUUCGUUAGGUGAUUUGGUUAUGCAGCAACAUCAUUAAAUCACCAAUUACUUUAGUCAACAGAGAAAUAAACUGAAUUUCCUUAUUC ..(.((((((((......))).(((....)))..(((((((((((.(((......))).))))))))))).........))))).)((.(((((.......)))))))..... ( -24.40, z-score = -1.42, R) >droGri2.scaffold_15110 6634533 111 + 24565398 --GUCACGUGGUUGUGCCAACUUCGAGUUCGUUAGGUGAUUUGGUUAUGCAACAACAUCAUUAAAUCACCAAUUACCUUAGUCAACAGAGGAAUAAACUGAAUUUCCUUAUAC --....((.(((((...))))).))((((((((.(((((((((((.(((......))).))))))))))).....((((.(....).))))....))).)))))......... ( -25.80, z-score = -1.68, R) >droWil1.scaffold_180698 150951 92 - 11422946 ---------------------UGCGACUUCGUUAGGUGAUUUGGUUAUGCAACAACAUCAUUAAAUCACCAAUUACUUUAGUCAACAGAGAAAUAAACCAAAUUUCCUUAUAC ---------------------...((((..((..(((((((((((.(((......))).)))))))))))....))...))))...((.(((((.......)))))))..... ( -20.00, z-score = -3.38, R) >consensus _____________________UGCGAGUUCGUUAGGUGAUUUGGUUAUGCAACAACAUCAUUAAAUCACCAAUUACUUUAGUCAACAGAGGAAUAAACCGAAUUUCCUUAUAC ..................................(((((((((((.(((......))).))))))))))).................((((((..........)))))).... (-18.34 = -18.59 + 0.25)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:44:10 2011