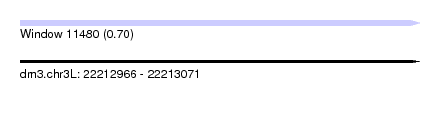
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 22,212,966 – 22,213,071 |
| Length | 105 |
| Max. P | 0.704381 |

| Location | 22,212,966 – 22,213,071 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 67.05 |
| Shannon entropy | 0.58174 |
| G+C content | 0.47331 |
| Mean single sequence MFE | -25.18 |
| Consensus MFE | -11.77 |
| Energy contribution | -11.60 |
| Covariance contribution | -0.17 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.22 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.704381 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
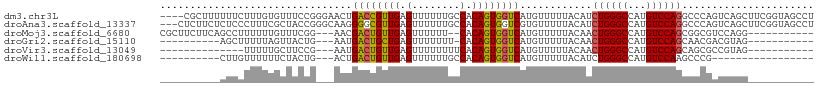
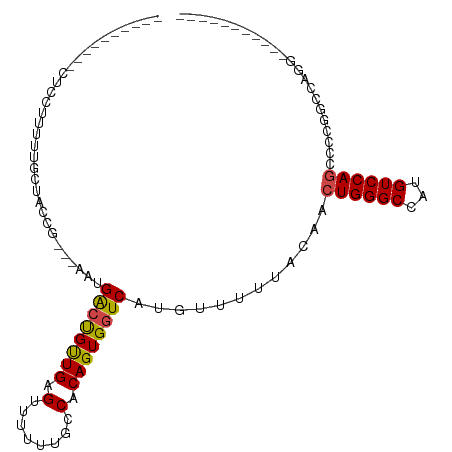
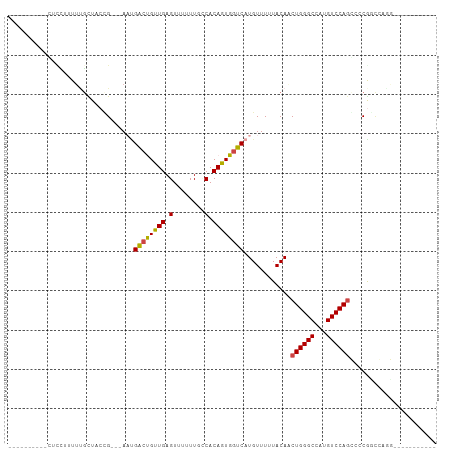
>dm3.chr3L 22212966 105 + 24543557 ----CGCUUUUUUCUUUGUGUUUCCGGGAACUGACCGUUGAGUUUUUUGCCACAGUGGUCAUGUUUUUACAUCUGGGCCAUGUCCAGGCCCAGUCAGCUUCGGUAGCCU ----...............(((.(((((((((((((((((.((.....))..))))))))).))))......(((((((.......)))))))......)))).))).. ( -30.80, z-score = -1.33, R) >droAna3.scaffold_13337 21373078 106 + 23293914 ---CUCUUCUCUCCCUUUCGCUACCGGGCAAGGGGCGUUGAGUUUUUUGCCACAGUGGUCGUGUUUUUACAUCUGGGCCAUGUCCAGGCCCAGUCAGCUUCGGUAGCCU ---................(((((((((((((((((.....))))))))))..(((....((((....))))(((((((.......)))))))...))).))))))).. ( -40.20, z-score = -2.89, R) >droMoj3.scaffold_6680 19557837 93 + 24764193 CGCUUCUUCAGCCUUUUUUGUUUCGG---AACGACUGUUGAGUUUUUU--CACAGUGGUCAUGUUUUUACAACUGGGCCAUGUCCAGCGGCGUCCAGG----------- ..........(((....((((..((.---.((.(((((.(((....))--)))))).))..)).....))))((((((...)))))).))).......----------- ( -23.50, z-score = -0.69, R) >droGri2.scaffold_15110 6621146 84 - 24565398 ----------AGCUUUUUAGUUACUG---AAUGACUGCUGAGUUUUUUU-CACAGUGGUCAUGUUUUUACAACUGGGCCAUGUCCAGCAACGACGUAG----------- ----------.........(((..((---.(((((..(((.(.......-).)))..)))))..........((((((...))))))))..)))....----------- ( -20.40, z-score = -0.89, R) >droVir3.scaffold_13049 14778587 81 + 25233164 --------------UUUUUGCUUCCG---AAUGACUGUUGAGUUUUUUUUCACAGUGGUCAUGUUUUUACAACUGGGCCAUGUCCAGCAGCGCCGUAG----------- --------------.....(((....---.(((((..(((.(........).)))..)))))..........((((((...)))))).))).......----------- ( -18.80, z-score = -0.71, R) >droWil1.scaffold_180698 135932 79 + 11422946 ----------CUUGUUUUUUCUACUG---ACUGACUGUUGAGUUUUUUGCCACAGUGGUCAUGUUUUUACAUCUGGGCCAUGUCCAAGCCCG----------------- ----------..(((.......((((---(((.(((((.(.((.....))))))))))))).))....)))...((((.........)))).----------------- ( -17.40, z-score = -0.80, R) >consensus __________CUCCUUUUUGCUACCG___AAUGACUGUUGAGUUUUUUGCCACAGUGGUCAUGUUUUUACAACUGGGCCAUGUCCAGCCCCGGCCAGG___________ ................................((((((((.(........).))))))))............((((((...))))))...................... (-11.77 = -11.60 + -0.17)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:44:07 2011