
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 22,198,449 – 22,198,552 |
| Length | 103 |
| Max. P | 0.986913 |

| Location | 22,198,449 – 22,198,552 |
|---|---|
| Length | 103 |
| Sequences | 7 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 59.59 |
| Shannon entropy | 0.79114 |
| G+C content | 0.48048 |
| Mean single sequence MFE | -17.56 |
| Consensus MFE | -6.88 |
| Energy contribution | -6.93 |
| Covariance contribution | 0.05 |
| Combinations/Pair | 1.58 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.39 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.26 |
| SVM RNA-class probability | 0.986913 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
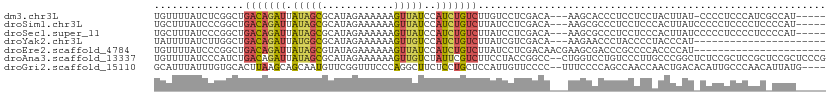
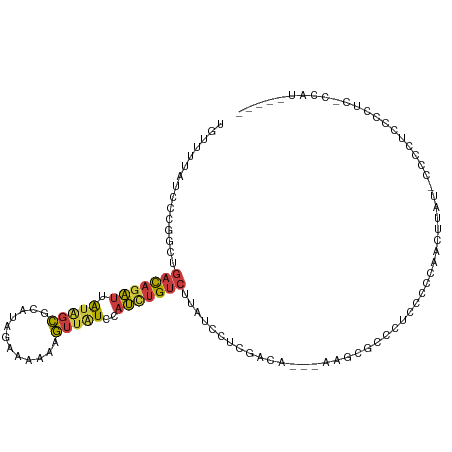
>dm3.chr3L 22198449 103 - 24543557 UGUUUUAUCUCGGCUGACAGAUUAUAGCGCAUAGAAAAAAGUUAUCCAUCUGUCUUGUCCUCGACA---AAGCACCCUCCUCCUACUUAU-CCCCUCCCAUCGCCAU----- ...........(((.(((((((.(((((............)))))..)))))))(((((...))))---)....................-...........)))..----- ( -18.70, z-score = -2.54, R) >droSim1.chr3L 21549389 104 - 22553184 UGCUUUAUCCCGGCUGACAGAUUAUAGCGCAUAGAAAAAAGUUAUCCAUCUGUCUUAUCCUCGACA---AAGCGCCCUCCUCCCACUUAUCCCCCUCCCCUCCCCAU----- .(((((.((..((..(((((((.(((((............)))))..)))))))....))..)).)---))))..................................----- ( -18.80, z-score = -3.27, R) >droSec1.super_11 2128830 104 - 2888827 UGCUUUAUCCCGGCUGACAGAUUAUAGCGCAUAGAAAAAAGUUAUCCAUCUGUCUUAUCCUCGACA---AAGCGCCCUCCUCCCACUUAUCCCCCUCCCCUCCCCAU----- .(((((.((..((..(((((((.(((((............)))))..)))))))....))..)).)---))))..................................----- ( -18.80, z-score = -3.27, R) >droYak2.chr3L 22583587 87 - 24197627 UAUUUUAUCUUGGCUGACAGAUUAUGGCGCAUAGAAAAAAGUUGUCCAUCUGUCUUAUCGUCGACA---AAGAACCCUACCCCUACCCAU---------------------- .........(((((.((((((...(((.(((...........)))))))))))).....)))))..---.....................---------------------- ( -12.70, z-score = -0.02, R) >droEre2.scaffold_4784 21869710 90 - 25762168 UGUUUUAUCCCGGCUGACAGAUUAUAGCGUAUAGAAAAAAGUUAUCCAUCUGUCUUAUCCUCGACAACGAAGCGACCCGCCCCACCCCAU---------------------- ...........(((.(((((((.(((((............)))))..)))))))...((((((....)).)).))...))).........---------------------- ( -16.30, z-score = -1.88, R) >droAna3.scaffold_13337 21360057 110 - 23293914 UGUUUUAUCCCAUCUGACAGAUUAUAGCGCAUAGAAAAAAGUUGUCUAUUCGUCUUCCUACCGGCC--CUGGUCCUGUCCCUUGCCCGGCUCUCCGCUCCGCUCCGCUCCCG ...............(((((......(((.(((((.........))))).))).........(((.--...))))))))....((..(((.....)))..)).......... ( -17.00, z-score = 0.86, R) >droGri2.scaffold_15110 6605149 106 + 24565398 GCAUUUAUUUGUGCACUUAAGCAGCAAUGUUCGGUUUCCCAGGCUUCUCCUGCUCCAUUGUUCCCC--UUUCCCCAGCCAACCAACUGACACAUUGCCCAACAUUAUG---- ((((......)))).........(((((((((((((.....((((.....................--.......))))....)))))).)))))))...........---- ( -20.59, z-score = -1.96, R) >consensus UGUUUUAUCCCGGCUGACAGAUUAUAGCGCAUAGAAAAAAGUUAUCCAUCUGUCUUAUCCUCGACA___AAGCGCCCUCCCCCAACUUAU_CCCCUCCCCUC_CCAU_____ ...............(((((((.(((((............)))))..))))))).......................................................... ( -6.88 = -6.93 + 0.05)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:44:04 2011