| Sequence ID | dm3.chr3L |
|---|---|
| Location | 22,192,161 – 22,192,263 |
| Length | 102 |
| Max. P | 0.765715 |

| Location | 22,192,161 – 22,192,263 |
|---|---|
| Length | 102 |
| Sequences | 10 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 76.45 |
| Shannon entropy | 0.44717 |
| G+C content | 0.48277 |
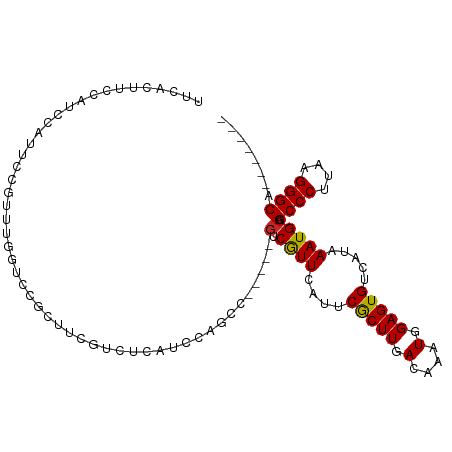
| Mean single sequence MFE | -23.79 |
| Consensus MFE | -12.16 |
| Energy contribution | -12.01 |
| Covariance contribution | -0.15 |
| Combinations/Pair | 1.13 |
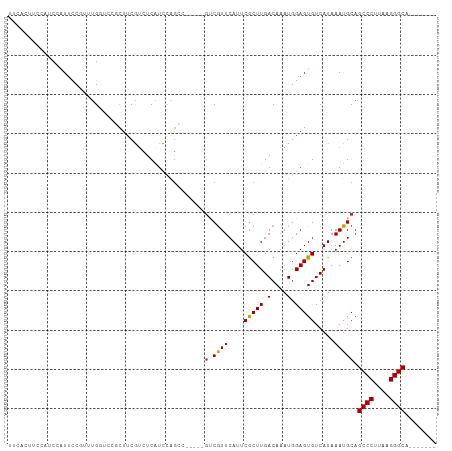
| Mean z-score | -1.79 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.765715 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
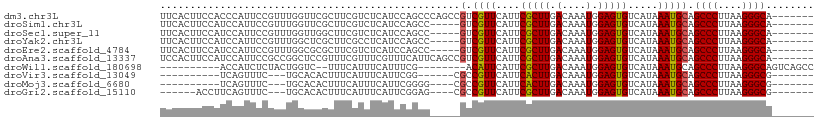
>dm3.chr3L 22192161 102 + 24543557 UUCACUUCCACCCAUUCCGUUUGGUUCGCUUCGUCUCAUCCAGCCCAGCCGUCGUUCAUUCGCUUGACAAAUGGAGUGUCAUAAAUGCAGCCCUUAAGGGCA------- ............((((((((((((((.(((...........)))..))))((((..(....)..))))))))))))))...........((((....)))).------- ( -26.10, z-score = -2.07, R) >droSim1.chr3L 21542286 97 + 22553184 UUCACUUCCAUCCAUUCCGUUUGGUUCGCUUCGUCUCAUCCAGCC-----GUCGUUCAUUCGCUUGACAAAUGGAGUGUCAUAAAUGCAGCCCUUAAGGGCA------- ............(((((((((((....((..((.((.....)).)-----)..))(((......))))))))))))))...........((((....)))).------- ( -23.60, z-score = -1.89, R) >droSec1.super_11 2122547 97 + 2888827 UUCACUUCCAUCCAUUCCGUUUGGUUGGCUUCGUCUCAUCCAGCC-----GUCGUUCAUUCGCUUGACAAAUGGAGUGUCAUAAAUGCAGCCCUUAAGGGCA------- ............((((((((((((((((...........))))))-----((((..(....)..))))))))))))))...........((((....)))).------- ( -29.70, z-score = -3.34, R) >droYak2.chr3L 22576575 97 + 24197627 UUCACUUCCAUCCAUUCCGUUUGGCUCGCUUCGCCUCAUCCAGCC-----GUCGUUCAUUCGCUUGACAAAUGGAGUGUCAUAAAUGCAGCCCUUAAGGGCA------- ............((((((((((((((...............))))-----((((..(....)..))))))))))))))...........((((....)))).------- ( -25.96, z-score = -2.08, R) >droEre2.scaffold_4784 21863728 97 + 25762168 UUCACUUCCAUCCAUUCCGUUUGGCGCGCUUCGUCUCAUCCAGCC-----GUCGUUCAUUCGCUUGACAAAUGGAGUGUCAUAAAUGCAGCCCUUAAGGGCA------- ............((((((((((((((.(((...........))))-----))...(((......))))))))))))))...........((((....)))).------- ( -26.40, z-score = -1.99, R) >droAna3.scaffold_13337 21354572 102 + 23293914 UCCACUUCCAUCCAUUCCGCCGGCUCCGUUUCGUUUCGUUUCAUUCAGCCGUCGUUCAUUCGCUUGACAAAUGGAGUGUCAUAAAUGCAGCCCUUAAGGGCA------- ..((((.((((......((.(((((......((...))........))))).)).(((......)))...))))))))...........((((....)))).------- ( -23.04, z-score = -1.21, R) >droWil1.scaffold_180698 108307 89 + 11422946 ----------ACCAUCUCUACUGGUC--UUUCAUUUCAUUUCG--------ACAUUCAUUCGCUUGACAAAUGGAGUGUCAUAAAUGCAGCCCUUAAGGGCAGUCAGCC ----------..........((((.(--(.......(((((.(--------(((((((((.(.....).))).)))))))..)))))..((((....)))))))))).. ( -19.00, z-score = -0.96, R) >droVir3.scaffold_13049 14755415 83 + 25233164 ----------UCAGUUUC---UGCACACUUUCAUUUCAUUCGG------CGCCGUUCAUUCACUUGACAAAUGGAGUGUCAUAAAUGCAGCCCUUAAGGGCG------- ----------........---((((......(((((((((((.------...)).(((......)))..))))))))).......))))((((....)))).------- ( -19.82, z-score = -1.48, R) >droMoj3.scaffold_6680 19535704 85 + 24764193 ----------UCAGUUUC---UGCACACUUUCAUUUCAUUCGGGG----CGCCGUUCAUUCACUUGACAAAUGGAGUGUCAUAAAUGCAGCCCUUAAGGGCG------- ----------........---((((......((((((((((((..----..))).(((......)))..))))))))).......))))((((....)))).------- ( -21.62, z-score = -1.44, R) >droGri2.scaffold_15110 6598930 89 - 24565398 ------ACCUUCAGUUUC---UGCACACUUUCAUUUCAUUCGGAG----CGCCGUUCAUUCGCUUGACAAAUGGAGUGUCAUAAAUGCAGCCCUUAAGGGCG------- ------............---((((......(((((((((.((((----((.........)))))..).))))))))).......))))((((....)))).------- ( -22.62, z-score = -1.39, R) >consensus UUCACUUCCAUCCAUUCCGUUUGGUCCGCUUCGUCUCAUCCAGCC_____GUCGUUCAUUCGCUUGACAAAUGGAGUGUCAUAAAUGCAGCCCUUAAGGGCA_______ ..................................................(.((((....(((((.(....).))))).....))))).((((....))))........ (-12.16 = -12.01 + -0.15)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:44:02 2011