
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 22,158,722 – 22,158,881 |
| Length | 159 |
| Max. P | 0.809892 |

| Location | 22,158,722 – 22,158,816 |
|---|---|
| Length | 94 |
| Sequences | 10 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 66.65 |
| Shannon entropy | 0.67898 |
| G+C content | 0.52654 |
| Mean single sequence MFE | -32.27 |
| Consensus MFE | -13.34 |
| Energy contribution | -12.01 |
| Covariance contribution | -1.33 |
| Combinations/Pair | 1.47 |
| Mean z-score | -1.06 |
| Structure conservation index | 0.41 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.658424 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
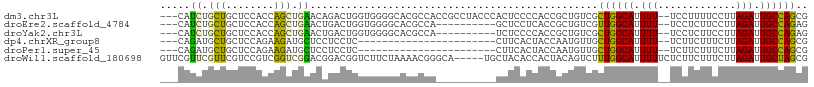
>dm3.chr3L 22158722 94 - 24543557 UGUCGCUGGCAUUUUUCCUUUUCCUUAGAUUGCCAGCGACAUUUCGACUGAGCUGCAGGAUUAUUGCG--GCGGUGGCGGUGGCGCUGGCGUUCUC----------- (((((((((((.(((...........))).)))))))))))....(((...(((((((.....)))))--))(.(((((....))))).))))...----------- ( -37.40, z-score = -2.18, R) >droEre2.scaffold_4784 21829762 91 - 25762168 UGUCGUUGGCAUUUUUCCUCUUCCUUAGAUUGCCAGAGACAUUUCGACUGAGCUGCAGGAUUAUUGCG--GCGGUGGCGGUGGCGGUAGCUUC-------------- ((((.((((((.(((...........))).)))))).)))).....((((.(((((((.....)))))--)).....))))(((....)))..-------------- ( -26.80, z-score = 0.20, R) >droYak2.chr3L 22513659 85 - 24197627 UGUCGCUGGCAUUUUUCCUCUUCCUUAGAUUGCCAGAGACAUUUCGACUCAGCUGCAGGAUUAUUGCG--GUGCUAGCGGUGGCUUC-------------------- ((((.((((((.(((...........))).)))))).))))..(((.((..(((((((.....)))))--))...))))).......-------------------- ( -25.50, z-score = -0.65, R) >droSec1.super_11 2089279 94 - 2888827 UGUCGCUGGCAUUUUUCCUCUUCCUUAGAUUGCCAGCGACAUUUCGACUGAGCUGCAGGAUUAUUGCG--GCGGUGGCGGUGGUGCUGGCGUUCUC----------- (((((((((((.(((...........))).)))))))))))........((((..(((.((((((((.--......)))))))).)))..))))..----------- ( -36.00, z-score = -1.80, R) >droSim1.chr3L 21507420 94 - 22553184 UGUCGCUGGCAUUUUUCCUCUUCCUUAGAUUGCCAGCGACAUUUCGACUGAGCUGCAGGAUUAUUGCG--GCGGUGGCGGUGGUGCUGGCGUUCUC----------- (((((((((((.(((...........))).)))))))))))........((((..(((.((((((((.--......)))))))).)))..))))..----------- ( -36.00, z-score = -1.80, R) >dp4.chrXR_group8 6234253 105 - 9212921 UGUUGCUGGCAUUUUUCUUCUUUCUUAGAUUGCCAGCGACAUUUCGACUGAGUUGCAGGAUUAUUGUG-CGGCAUGGCUGAGUGUGUGGCACAGGCACGGUCACGG- (((((((((((....(((........))).)))))))))))....(((((.((((((.((...)).))-))))...(((..(((.....))).))).)))))....- ( -40.60, z-score = -2.91, R) >droPer1.super_45 369889 105 + 618639 UGUUGCUGGCAUUUUUCUUCUUUCUUAGAUUGCCAGCGACAUUUCGACUGAGUUGCAGGAUUAUUGUG-CGGCAUGGCUGAGUGUGUGGCACAGGCACGGUCACGG- (((((((((((....(((........))).)))))))))))....(((((.((((((.((...)).))-))))...(((..(((.....))).))).)))))....- ( -40.60, z-score = -2.91, R) >droMoj3.scaffold_6680 19489422 94 - 24764193 UGCCAUUGGCGUUUUUCUU-------AGAUUGCUGGCGACGUUCCG-UUGAGUUGCAGGAUUAUUGUGUUGGGGCAGUCA-GACG----GAGGGGGUUGACAAGUGU .((((...(((.(((....-------))).)))))))..(.(((((-(((((((.(((.((....)).))).)))..)))-.)))----))).)............. ( -22.70, z-score = 0.91, R) >droVir3.scaffold_13049 14714799 95 - 25233164 UGCCAUUGGCGUUUUUCUU-------AGAUUGCUGGCGACGUUCCG-UUGAGUUGCAGGAUUAUUGUGUUGGGGCAGUCGCGAGG----CGCGGGGCUGACAAGUGU .((((...(((.(((....-------))).)))))))...((((((-(...(((.(((.((....)).))).))).(((....))----)))))))).......... ( -27.10, z-score = 1.06, R) >droGri2.scaffold_15110 6559368 101 + 24565398 UGCCUUUGGCGUUUUUUCUUC----CAGAUUGCUGGUGGAGUUCCG-UUGAGUUGCAGGAUUAUUGUAUUGGGGCAGUCGCGUCGCCUCGUAGAGGAUGACAAGUG- ((((((..(((....((((.(----(((....)))).))))...))-)..)..(((((.....)))))...))))).....((((((((...)))).)))).....- ( -30.00, z-score = -0.54, R) >consensus UGUCGCUGGCAUUUUUCCUCUUCCUUAGAUUGCCAGCGACAUUUCGACUGAGCUGCAGGAUUAUUGCG__GGGGUGGCGGUGGCGCUGGCGUGGGC___________ ((((.((((((.(((...........))).)))))).)))).............((((.....))))........................................ (-13.34 = -12.01 + -1.33)

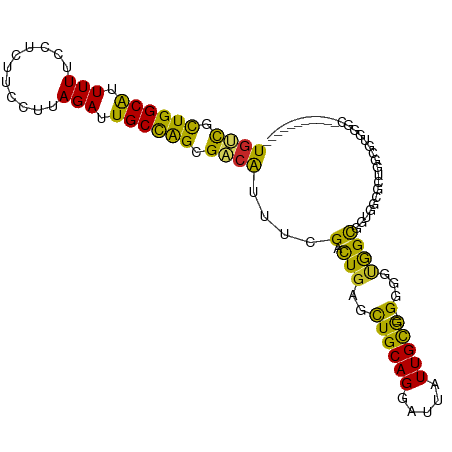

| Location | 22,158,748 – 22,158,848 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 75.62 |
| Shannon entropy | 0.41370 |
| G+C content | 0.52655 |
| Mean single sequence MFE | -28.28 |
| Consensus MFE | -15.15 |
| Energy contribution | -15.73 |
| Covariance contribution | 0.58 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.660765 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 22158748 100 + 24543557 ---------CGCAAUAAUCCUGCAGCU-CAGUCGAAAUGUCGCUGGCAAUCUAAGGAAAAGGA--AAAAUGCCAGCGACAGCGGUGGGG-AGUGGGUAGGCGGUGGCGUGCCC ---------.(((.......))).(((-(.((((...(((((((((((.(((........)))--....))))))))))).))))...)-)))(((((.((....)).))))) ( -40.50, z-score = -3.17, R) >droEre2.scaffold_4784 21829785 90 + 25762168 ---------CGCAAUAAUCCUGCAGCU-CAGUCGAAAUGUCUCUGGCAAUCUAAGGAAGAGGA--AAAAUGCCAACGACAGCGGUGAGG-AGCUGGCGUGCCC---------- ---------.(((.....((.((..((-((.(((...((((..(((((.(((........)))--....)))))..)))).))))))).-.)).))..)))..---------- ( -25.90, z-score = -0.44, R) >droYak2.chr3L 22513676 90 + 24197627 ---------CGCAAUAAUCCUGCAGCU-GAGUCGAAAUGUCUCUGGCAAUCUAAGGAAGAGGA--AAAAUGCCAGCGACAGCGGUGGGG-AGAUGGCGUGCCC---------- ---------(((.....((((...(((-(........((((.((((((.(((........)))--....)))))).)))).)))).)))-)....))).....---------- ( -26.30, z-score = -0.25, R) >droSec1.super_11 2089305 77 + 2888827 ---------CGCAAUAAUCCUGCAGCU-CAGUCGAAAUGUCGCUGGCAAUCUAAGGAAGAGGA--AAAAUGCCAGCGACAGCGGUGGGG------------------------ ---------.(((.......)))..((-((.(((...(((((((((((.(((........)))--....))))))))))).))))))).------------------------ ( -29.80, z-score = -3.56, R) >droSim1.chr3L 21507446 77 + 22553184 ---------CGCAAUAAUCCUGCAGCU-CAGUCGAAAUGUCGCUGGCAAUCUAAGGAAGAGGA--AAAAUGCCAGCGACAGCGGUGGGG------------------------ ---------.(((.......)))..((-((.(((...(((((((((((.(((........)))--....))))))))))).))))))).------------------------ ( -29.80, z-score = -3.56, R) >droWil1.scaffold_180698 11355711 103 - 11422946 CUGCCCCAACACAAUAAUCCUGCAACUUCAGGCAAAACGUCGCUAGCAAUCUAAGAAAGAAGAGAAAAAUGCCAAAGACUGUAGUGGUGUAGCAUGCCCGUUU---------- ((((.(((..(((.....((((......))))......(((....(((.(((........)))......)))....))))))..))).))))...........---------- ( -17.40, z-score = 0.70, R) >consensus _________CGCAAUAAUCCUGCAGCU_CAGUCGAAAUGUCGCUGGCAAUCUAAGGAAGAGGA__AAAAUGCCAGCGACAGCGGUGGGG_AG__GGC__GC____________ .................((((((.(((...........(((.((((((.(((........)))......)))))).)))))).))))))........................ (-15.15 = -15.73 + 0.58)

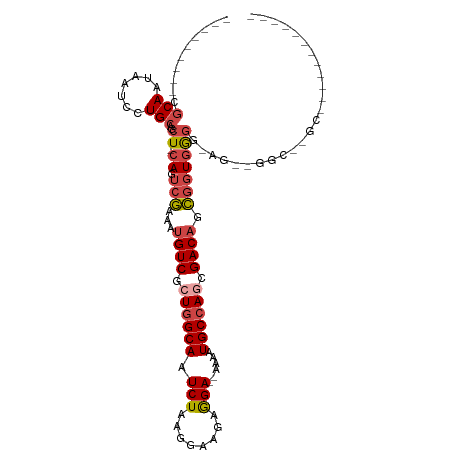
| Location | 22,158,748 – 22,158,848 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 75.62 |
| Shannon entropy | 0.41370 |
| G+C content | 0.52655 |
| Mean single sequence MFE | -24.70 |
| Consensus MFE | -15.33 |
| Energy contribution | -14.72 |
| Covariance contribution | -0.61 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.809892 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 22158748 100 - 24543557 GGGCACGCCACCGCCUACCCACU-CCCCACCGCUGUCGCUGGCAUUUU--UCCUUUUCCUUAGAUUGCCAGCGACAUUUCGACUG-AGCUGCAGGAUUAUUGCG--------- (((...((....))...))).((-(........(((((((((((.(((--...........))).)))))))))))........)-))..((((.....)))).--------- ( -30.19, z-score = -2.87, R) >droEre2.scaffold_4784 21829785 90 - 25762168 ----------GGGCACGCCAGCU-CCUCACCGCUGUCGUUGGCAUUUU--UCCUCUUCCUUAGAUUGCCAGAGACAUUUCGACUG-AGCUGCAGGAUUAUUGCG--------- ----------..(((..((.((.-.((((.((.((((.((((((.(((--...........))).)))))).))))...))..))-))..)).)).....))).--------- ( -23.80, z-score = -0.32, R) >droYak2.chr3L 22513676 90 - 24197627 ----------GGGCACGCCAUCU-CCCCACCGCUGUCGCUGGCAUUUU--UCCUCUUCCUUAGAUUGCCAGAGACAUUUCGACUC-AGCUGCAGGAUUAUUGCG--------- ----------(((..........-.)))..((.((((.((((((.(((--...........))).)))))).))))...))....-....((((.....)))).--------- ( -20.60, z-score = 0.07, R) >droSec1.super_11 2089305 77 - 2888827 ------------------------CCCCACCGCUGUCGCUGGCAUUUU--UCCUCUUCCUUAGAUUGCCAGCGACAUUUCGACUG-AGCUGCAGGAUUAUUGCG--------- ------------------------......((((((((((((((.(((--...........))).)))))))))))......(((-.....))).......)))--------- ( -25.00, z-score = -3.44, R) >droSim1.chr3L 21507446 77 - 22553184 ------------------------CCCCACCGCUGUCGCUGGCAUUUU--UCCUCUUCCUUAGAUUGCCAGCGACAUUUCGACUG-AGCUGCAGGAUUAUUGCG--------- ------------------------......((((((((((((((.(((--...........))).)))))))))))......(((-.....))).......)))--------- ( -25.00, z-score = -3.44, R) >droWil1.scaffold_180698 11355711 103 + 11422946 ----------AAACGGGCAUGCUACACCACUACAGUCUUUGGCAUUUUUCUCUUCUUUCUUAGAUUGCUAGCGACGUUUUGCCUGAAGUUGCAGGAUUAUUGUGUUGGGGCAG ----------.........(((..((.(((....(((.((((((.(((.............))).)))))).)))......((((......))))......))).))..))). ( -23.62, z-score = 0.36, R) >consensus ____________GC__GCC__CU_CCCCACCGCUGUCGCUGGCAUUUU__UCCUCUUCCUUAGAUUGCCAGCGACAUUUCGACUG_AGCUGCAGGAUUAUUGCG_________ ..............................(((.(((.((((((.(((.............))).)))))).))).......(((......))).......)))......... (-15.33 = -14.72 + -0.61)


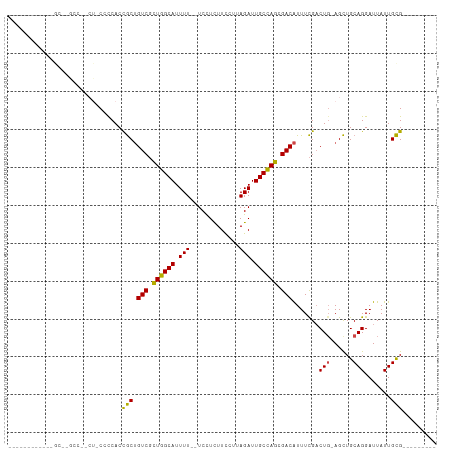
| Location | 22,158,778 – 22,158,881 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 65.27 |
| Shannon entropy | 0.60520 |
| G+C content | 0.53294 |
| Mean single sequence MFE | -27.64 |
| Consensus MFE | -7.35 |
| Energy contribution | -6.69 |
| Covariance contribution | -0.66 |
| Combinations/Pair | 1.57 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.27 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.662313 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 22158778 103 - 24543557 ---CAUCUGCUGCUCCACCAGCUGAACAGACUGGUGGGGCACGCCACCGCCUACCCACUCCCCACCGCUGUCGCUGGCAUUUU--UCCUUUUCCUUAGAUUGCCAGCG ---.....(((((((((((((((....)).))))))))))).))...........................((((((((.(((--...........))).)))))))) ( -37.80, z-score = -4.23, R) >droEre2.scaffold_4784 21829815 93 - 25762168 ---CAUCUGCUGCUCCACCAGCUGAACUGACUGGUGGGGCACGCCA----------GCUCCUCACCGCUGUCGUUGGCAUUUU--UCCUCUUCCUUAGAUUGCCAGAG ---.....((((((((((((((......).))))))))))).))((----------((........))))...((((((.(((--...........))).)))))).. ( -34.10, z-score = -2.69, R) >droYak2.chr3L 22513706 93 - 24197627 ---CAUCUGCUGCUCCACCAGCUGAACUGACUGGUGGGGCACGCCA----------UCUCCCCACCGCUGUCGCUGGCAUUUU--UCCUCUUCCUUAGAUUGCCAGAG ---.....((((((((((((((......).))))))))))).))..----------.................((((((.(((--...........))).)))))).. ( -32.10, z-score = -2.57, R) >dp4.chrXR_group8 6234320 80 - 9212921 ---CAGAUGCUGCUCCAGAAGAUGCUCCUCCUC-----------------------CUUCACUACCAAUGUUGCUGGCAUUUU--UCUUCUUUCUUAGAUUGCCAGCG ---..............((((..(......)..-----------------------))))............(((((((....--(((........))).))))))). ( -16.80, z-score = -0.80, R) >droPer1.super_45 369956 80 + 618639 ---CAGAUGCUGCUCCAGAAGAUGCUCCUCCUC-----------------------CUUCACUACCAAUGUUGCUGGCAUUUU--UCUUCUUUCUUAGAUUGCCAGCG ---..............((((..(......)..-----------------------))))............(((((((....--(((........))).))))))). ( -16.80, z-score = -0.80, R) >droWil1.scaffold_180698 11355751 103 + 11422946 GUUCGUUCGUUCGUCCGUCGGUCGGACGGACGGUCUUCUAAAACGGGCA-----UGCUACACCACUACAGUCUUUGGCAUUUUUCUCUUCUUUCUUAGAUUGCUAGCG (((((((((((((((((.....))))))))))).........)))))).-----..........(((((((((..((...............))..)))))).))).. ( -28.26, z-score = -2.12, R) >consensus ___CAUCUGCUGCUCCACCAGCUGAACCGACUGGUGGGGCACGCCA__________CCUCACCACCACUGUCGCUGGCAUUUU__UCCUCUUCCUUAGAUUGCCAGCG .....((.(((........))).))................................................((((((.(((.............))).)))))).. ( -7.35 = -6.69 + -0.66)
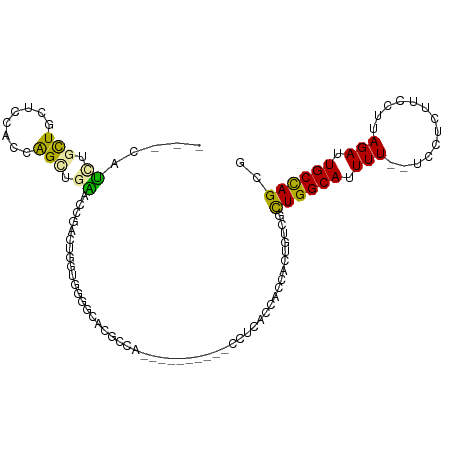

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:43:58 2011