
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 22,154,001 – 22,154,093 |
| Length | 92 |
| Max. P | 0.824675 |

| Location | 22,154,001 – 22,154,093 |
|---|---|
| Length | 92 |
| Sequences | 9 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 68.29 |
| Shannon entropy | 0.57770 |
| G+C content | 0.45036 |
| Mean single sequence MFE | -12.88 |
| Consensus MFE | -6.60 |
| Energy contribution | -6.82 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.23 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.81 |
| SVM RNA-class probability | 0.824675 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 22154001 92 - 24543557 ------UUCCCCGUUC----UCGAUCUCAUUCGCAUUCCCAG------------UCCGGACU--CCCGUCCCGUGCCCAUUCCUAAUGCCAUAAAUAAAAAUCAGUUUAUGUGCGG- ------....((((..----............(((((....(------------..(((((.--...)).)))..)........)))))(((((((........))))))).))))- ( -14.60, z-score = -1.62, R) >droPer1.super_45 363757 89 + 618639 ------UUCCCCAUUCCC--CCAUUCCCAGUCGCAUUUCCAU------------UUC-------CAUUUCCAUGGCCCCUUGCUAAUGCCAUAAAUAAAAAUCAGUUUAUGUGCGG- ------............--......((....(((...((((------------...-------.......)))).....)))....(((((((((........))))))).))))- ( -12.00, z-score = -1.56, R) >dp4.chrXR_group8 6228061 89 - 9212921 ------UUCCCCAUUCCC--CCAUUCCCAGUCGCAUUUCCAU------------UUC-------CAUUUCCAUGGCCCCUUGCUAAUGCCAUAAAUAAAAAUCAGUUUAUGUGCGG- ------............--......((....(((...((((------------...-------.......)))).....)))....(((((((((........))))))).))))- ( -12.00, z-score = -1.56, R) >droAna3.scaffold_13337 21320106 116 - 23293914 UUCCUCGUUCUCAUUCCCCGUCGAUCUCAUUCGCAUUCCCAGAGCCAUCUUAUAUCCCAUCUACCACAUCCCAGGCCCAUUGCUAAUGCCAUAAAUAAAAAUCAGUUUAUGUGCGG- .................((((...........(((((..(((.(((...........................)))...)))..)))))(((((((........))))))).))))- ( -14.13, z-score = -0.76, R) >droEre2.scaffold_4784 21825176 74 - 25762168 ----------------------------UUCCCCAUUCCCAG------------UCCAGAAU--CCCGUCCCAUGCCCAUUGCUAAUGCCAUAAAUAAAAAUCAGUUUAUGUGCGG- ----------------------------..............------------........--......((..((.....))....(((((((((........))))))).))))- ( -8.00, z-score = -0.77, R) >droYak2.chr3L 22508958 92 - 24197627 ------UUCCCCAUUC----UCGAUCACAUUCGCAUUCUCAG------------UCCAGGCU--CCCGUCCCAUGCCCAUUGCUAAUGCCAUAAAUAAAAAUCAGUUUAUGUGCGG- ------....((....----............(((((..(((------------(...(((.--..........))).))))..)))))(((((((........)))))))...))- ( -14.30, z-score = -1.20, R) >droSec1.super_11 2084060 92 - 2888827 ------UUCCCCAUUC----UCGAUCUCAUUCGCAUUCACAG------------UCCGGACU--CCCGUCCCAUGCCCAUUGCUAAUGCCAUAAAUAAAAAUCAGUUUAUGUGCGG- ------....((....----............(((....((.------------...((((.--...))))..)).....)))....(((((((((........))))))).))))- ( -15.50, z-score = -1.57, R) >droSim1.chr3L 21502617 92 - 22553184 ------UUCCCCAUUC----UCGAUCUCAUUCGCAUUCCCAG------------UCCGGACU--CCCGUCCCAUGCCCAUUGCUAAUGCCAUAAAUAAAAAUCAGUUUAUGUGCGG- ------....((....----............(((....((.------------...((((.--...))))..)).....)))....(((((((((........))))))).))))- ( -15.50, z-score = -1.79, R) >droWil1.scaffold_180698 54581 80 - 11422946 -------------------------UUUCCCUAGAUAUAUAU------------ACUAUAUAUACAUAUAUGUUGUGUUCCCUCGAACCCAUAAAUAAAAAUCCGUUUAUGUGCGGC -------------------------........(((((((((------------...........)))))))))(.((((....)))).)............((((......)))). ( -9.90, z-score = -0.22, R) >consensus ______UUCCCCAUUC____UCGAUCUCAUUCGCAUUCCCAG____________UCCAGACU__CCCGUCCCAUGCCCAUUGCUAAUGCCAUAAAUAAAAAUCAGUUUAUGUGCGG_ ......................................................................................((((((((((........))))))).))).. ( -6.60 = -6.82 + 0.22)
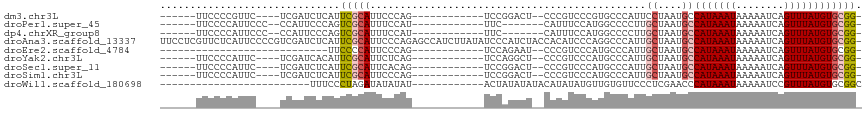
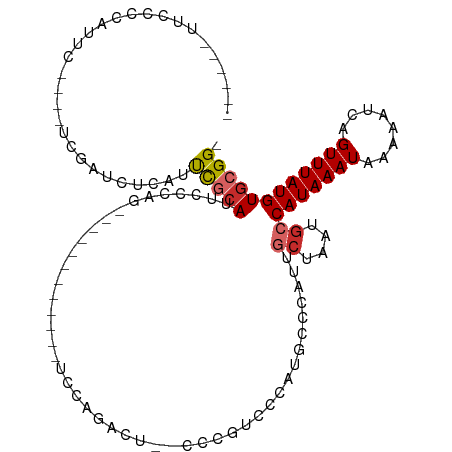
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:43:55 2011