| Sequence ID | dm3.chr3L |
|---|---|
| Location | 22,142,881 – 22,142,981 |
| Length | 100 |
| Max. P | 0.968689 |

| Location | 22,142,881 – 22,142,981 |
|---|---|
| Length | 100 |
| Sequences | 8 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 81.35 |
| Shannon entropy | 0.36489 |
| G+C content | 0.34446 |
| Mean single sequence MFE | -25.96 |
| Consensus MFE | -21.10 |
| Energy contribution | -20.52 |
| Covariance contribution | -0.58 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.81 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.80 |
| SVM RNA-class probability | 0.968689 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 22142881 100 - 24543557 ---UAUUAUUUGCAUUUGAAUCAUUCGUUUGUGCGAAAUCAGAUUAAAUCGCAUAAUCGAAUCUGAUGUUUAGUGUGUGGGGCCAUUCUACAUAUGAAAAAAA--------- ---........(((((.((....((((.((((((((............)))))))).)))))).)))))...(((((((((.....)))))))))........--------- ( -23.00, z-score = -1.54, R) >droSim1.chr3L 21490973 102 - 22553184 ---UGUUAUUUGCAUUUGAAUCAUUCGUUUGUGCGAAAUCAGAUUAAAUCGCAUAAUCGAAUCUGAUGCUUUGUGUGUGGGGCCAUUCUACAUAUGAAAAAAAAA------- ---........(((((.((....((((.((((((((............)))))))).)))))).)))))((..((((((((.....))))))))..)).......------- ( -27.20, z-score = -2.63, R) >droSec1.super_11 2072949 102 - 2888827 ---UGUUAUUUGCAUUUGAAUCAUUCGUUUGUGCGAAAUCAGAUUAAAUCGCAUAAUCGAAUCUGAUGCUUUGUGUGUGGGGCCAUUCUACAUAUGAAAAAAAAA------- ---........(((((.((....((((.((((((((............)))))))).)))))).)))))((..((((((((.....))))))))..)).......------- ( -27.20, z-score = -2.63, R) >droYak2.chr3L 22494341 100 - 24197627 ---UGUUAUUUGCAUUUGAAUCAUUCGUUUGUGCGAAAUCAGAUUAAAUCGCAUAAUCGAAUCUGAUGCUUUGUGUGUGGGGCCAUUCUACAUAUGAAAAAAA--------- ---........(((((.((....((((.((((((((............)))))))).)))))).)))))((..((((((((.....))))))))..)).....--------- ( -27.20, z-score = -2.56, R) >droEre2.scaffold_4784 21814303 106 - 25762168 ---UGUUAUUUGCAUUUGAAUCAUUCGUUUGUGCGAAAUCAGAUUAAAUCGCAUAAUCGAAUCUGAUGCUUUGUGUGUGGGGCCAUUCUACAUUUGAAAAAACGAAAAA--- ---.(((....(((((.((....((((.((((((((............)))))))).)))))).)))))((((.(((((((.....))))))).))))..)))......--- ( -23.70, z-score = -0.88, R) >droAna3.scaffold_13337 21309538 90 - 23293914 GCCUGGUAUUUGCAUUUGAAUCAUUCGUUUGUGCGAAAUCAGAUUAAGUCGCAUAAUCGAAUCUGGUGCUU--UAUGUGCCGCUCGCACACG-------------------- ((.((((((..((((..((....((((.((((((((............)))))))).))))))..))))..--...))))))...)).....-------------------- ( -24.70, z-score = -1.19, R) >dp4.chrXR_group8 6215154 108 - 9212921 ---UGUUAUUUGCAUUUGAAUCAUUCGUUUGUGCGAAAUCAGAUUAAAUCGCAUAAUCGAAUCUGAUGCUACGUAUAUGU-ACGACUAUAUAUAUGUAUAUGUAUAGAGUCG ---........(((((.((....((((.((((((((............)))))))).)))))).)))))...........-.(((((.(((((((....))))))).))))) ( -27.40, z-score = -1.62, R) >droPer1.super_45 349321 103 + 618639 ---UGUUAUUUGCAUUUGAAUCAUUCGUUUGUGCGAAAUCAGAUUAAAUCGCAUAAUCGAAUCUGAUGCUACGUAUAUGUGACGACUAUAUA------UAUGUAUAGAGUCG ---........(((((.((....((((.((((((((............)))))))).)))))).)))))((((((((((((.....))))))------))))))........ ( -27.30, z-score = -1.87, R) >consensus ___UGUUAUUUGCAUUUGAAUCAUUCGUUUGUGCGAAAUCAGAUUAAAUCGCAUAAUCGAAUCUGAUGCUUUGUGUGUGGGGCCAUUCUACAUAUGAAAAAAAA________ ...........(((((.((....((((.((((((((............)))))))).)))))).))))).....(((((((.....)))))))................... (-21.10 = -20.52 + -0.58)

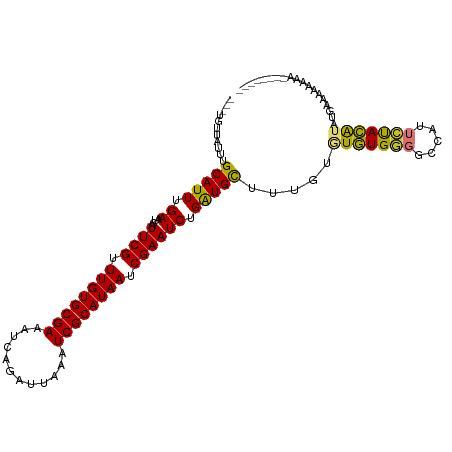
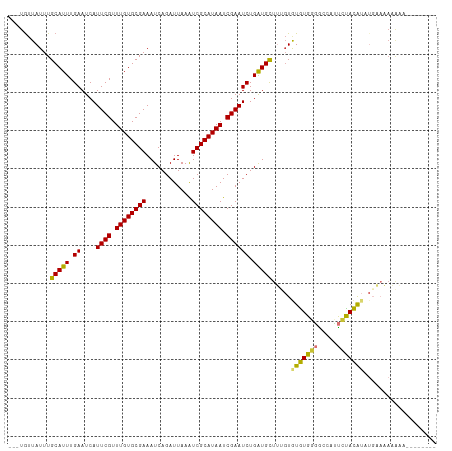
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:43:54 2011