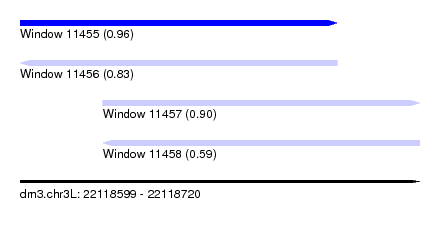
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 22,118,599 – 22,118,720 |
| Length | 121 |
| Max. P | 0.960084 |

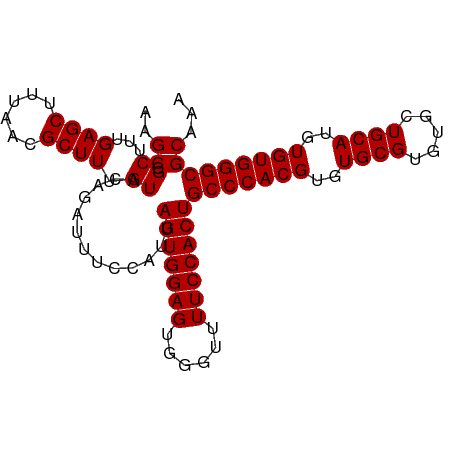
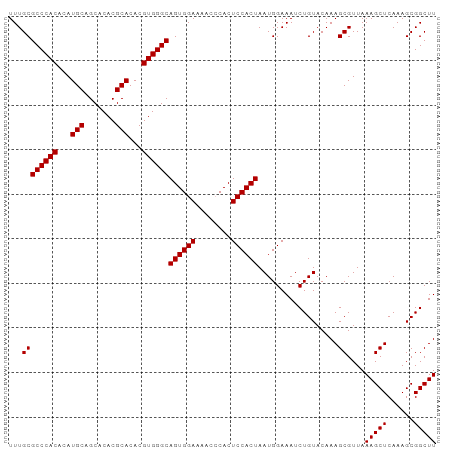
| Location | 22,118,599 – 22,118,695 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 99.38 |
| Shannon entropy | 0.01011 |
| G+C content | 0.50625 |
| Mean single sequence MFE | -35.10 |
| Consensus MFE | -35.10 |
| Energy contribution | -35.10 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.90 |
| Structure conservation index | 1.00 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.68 |
| SVM RNA-class probability | 0.960084 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 22118599 96 + 24543557 AAGCCGCUUUGAGCUUUAACGCUUUGUACAGAUUUCCAUUAGUGGAGUGGGUUUUCCACUGCCCACGUGUGCGUGUGCUGCAUGUGUGGGCGCAAA ..((.((...((((......)))).)).............(((((((......)))))))(((((((..((((.....))))..)))))))))... ( -35.10, z-score = -1.70, R) >droSim1.chr3L 21465632 96 + 22553184 AAGCCGCUUUGAGCUUUAACGCUUUGUACAGAUUUCCAUUAGUGGAGUGGGUUUUCCACUGCCCACGUGUGCGUGUGCUGCAUGUGUGGGCGCAAA ..((.((...((((......)))).)).............(((((((......)))))))(((((((..((((.....))))..)))))))))... ( -35.10, z-score = -1.70, R) >droSec1.super_11 2048007 96 + 2888827 AAGCCGCUUUGAGCUUUAACGCUUUGUACAGAUUUCCAUUAGUGGAGUGGGUUUUCCACUGCCCACGUGUGCGUGUGCUGCAUGUGUGGGCGCAAA ..((.((...((((......)))).)).............(((((((......)))))))(((((((..((((.....))))..)))))))))... ( -35.10, z-score = -1.70, R) >droYak2.chr3L 22461975 96 + 24197627 AAGCCGCUUUGAGCUUUAACGCUUUGUACAGAUUUCCAUUAGUGGAGUGUGUUUUCCACUGCCCACGUGUGCGUGUGCUGCAUGUGUGGGCGCAAA ..((.((...((((......)))).)).............(((((((......)))))))(((((((..((((.....))))..)))))))))... ( -35.10, z-score = -2.20, R) >droEre2.scaffold_4784 21789504 96 + 25762168 AAGCCGCUUUGAGCUUUAACGCUUUGUACAGAUUUCCAUUAGUGGAGUGUGUUUUCCACUGCCCACGUGUGCGUGUGCUGCAUGUGUGGGCGCAAA ..((.((...((((......)))).)).............(((((((......)))))))(((((((..((((.....))))..)))))))))... ( -35.10, z-score = -2.20, R) >consensus AAGCCGCUUUGAGCUUUAACGCUUUGUACAGAUUUCCAUUAGUGGAGUGGGUUUUCCACUGCCCACGUGUGCGUGUGCUGCAUGUGUGGGCGCAAA ..((.((...((((......)))).)).............(((((((......)))))))(((((((..((((.....))))..)))))))))... (-35.10 = -35.10 + 0.00)

| Location | 22,118,599 – 22,118,695 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 99.38 |
| Shannon entropy | 0.01011 |
| G+C content | 0.50625 |
| Mean single sequence MFE | -27.60 |
| Consensus MFE | -27.60 |
| Energy contribution | -27.60 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.31 |
| Structure conservation index | 1.00 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.83 |
| SVM RNA-class probability | 0.829145 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 22118599 96 - 24543557 UUUGCGCCCACACAUGCAGCACACGCACACGUGGGCAGUGGAAAACCCACUCCACUAAUGGAAAUCUGUACAAAGCGUUAAAGCUCAAAGCGGCUU ...((((((((...(((.......)))...))))))((((((........))))))..................))....(((((......))))) ( -27.60, z-score = -1.22, R) >droSim1.chr3L 21465632 96 - 22553184 UUUGCGCCCACACAUGCAGCACACGCACACGUGGGCAGUGGAAAACCCACUCCACUAAUGGAAAUCUGUACAAAGCGUUAAAGCUCAAAGCGGCUU ...((((((((...(((.......)))...))))))((((((........))))))..................))....(((((......))))) ( -27.60, z-score = -1.22, R) >droSec1.super_11 2048007 96 - 2888827 UUUGCGCCCACACAUGCAGCACACGCACACGUGGGCAGUGGAAAACCCACUCCACUAAUGGAAAUCUGUACAAAGCGUUAAAGCUCAAAGCGGCUU ...((((((((...(((.......)))...))))))((((((........))))))..................))....(((((......))))) ( -27.60, z-score = -1.22, R) >droYak2.chr3L 22461975 96 - 24197627 UUUGCGCCCACACAUGCAGCACACGCACACGUGGGCAGUGGAAAACACACUCCACUAAUGGAAAUCUGUACAAAGCGUUAAAGCUCAAAGCGGCUU ...((((((((...(((.......)))...))))))((((((........))))))..................))....(((((......))))) ( -27.60, z-score = -1.45, R) >droEre2.scaffold_4784 21789504 96 - 25762168 UUUGCGCCCACACAUGCAGCACACGCACACGUGGGCAGUGGAAAACACACUCCACUAAUGGAAAUCUGUACAAAGCGUUAAAGCUCAAAGCGGCUU ...((((((((...(((.......)))...))))))((((((........))))))..................))....(((((......))))) ( -27.60, z-score = -1.45, R) >consensus UUUGCGCCCACACAUGCAGCACACGCACACGUGGGCAGUGGAAAACCCACUCCACUAAUGGAAAUCUGUACAAAGCGUUAAAGCUCAAAGCGGCUU ...((((((((...(((.......)))...))))))((((((........))))))..................))....(((((......))))) (-27.60 = -27.60 + -0.00)

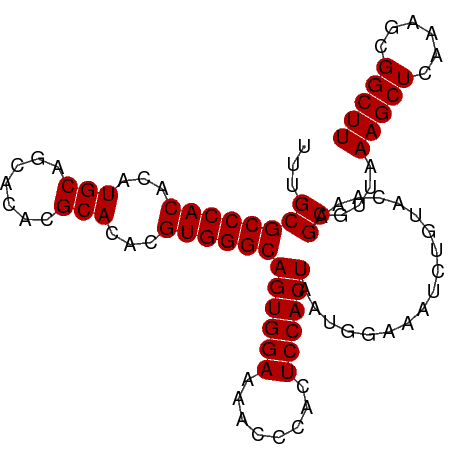
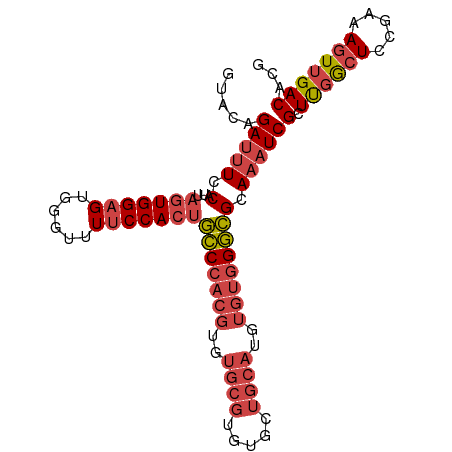
| Location | 22,118,624 – 22,118,720 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 84.10 |
| Shannon entropy | 0.30915 |
| G+C content | 0.54019 |
| Mean single sequence MFE | -34.88 |
| Consensus MFE | -26.91 |
| Energy contribution | -28.50 |
| Covariance contribution | 1.59 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.77 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.13 |
| SVM RNA-class probability | 0.896212 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 22118624 96 + 24543557 GUACAGAUUUCCAUUAGUGGAGUGGGUUUUCCACUGCCCACGUGUGCGUGUGCUGCAUGUGUGGGCGCAAAUCGCUUCGCUCCGAAAGUUGACACG ((.(((.((((....(((((((((((....))...(((((((..((((.....))))..)))))))......)))))))))..)))).)))))... ( -37.60, z-score = -2.41, R) >droSim1.chr3L 21465657 96 + 22553184 GUACAGAUUUCCAUUAGUGGAGUGGGUUUUCCACUGCCCACGUGUGCGUGUGCUGCAUGUGUGGGCGCAAAUCGCUUGGCUCCGAAAGUUGACACG .....(((((.(...(((((((......)))))))(((((((..((((.....))))..)))))))).)))))(.(..(((.....)))..))... ( -35.90, z-score = -1.64, R) >droSec1.super_11 2048032 96 + 2888827 GUACAGAUUUCCAUUAGUGGAGUGGGUUUUCCACUGCCCACGUGUGCGUGUGCUGCAUGUGUGGGCGCAAAUCGCUUGGCUCCGAAAGUUGACACG .....(((((.(...(((((((......)))))))(((((((..((((.....))))..)))))))).)))))(.(..(((.....)))..))... ( -35.90, z-score = -1.64, R) >droYak2.chr3L 22462000 96 + 24197627 GUACAGAUUUCCAUUAGUGGAGUGUGUUUUCCACUGCCCACGUGUGCGUGUGCUGCAUGUGUGGGCGCAAAUCGCUUGGCUCCGAAAGUUGACACG .....(((((.(...(((((((......)))))))(((((((..((((.....))))..)))))))).)))))(.(..(((.....)))..))... ( -35.90, z-score = -2.02, R) >droEre2.scaffold_4784 21789529 96 + 25762168 GUACAGAUUUCCAUUAGUGGAGUGUGUUUUCCACUGCCCACGUGUGCGUGUGCUGCAUGUGUGGGCGCAAAUCGCUCGGCUCCGAAAGUUGACACG .....(((((.(...(((((((......)))))))(((((((..((((.....))))..)))))))).)))))(.((((((.....)))))))... ( -37.30, z-score = -2.36, R) >droAna3.scaffold_13337 21283716 81 + 23293914 ----GGACUUGCCCGUGUGGGGCAAGUUUGCCAGUCUCCA--------UGUGCAACACGUGCAGAGGAAUCUCGCUCGACUCAGAAA-UUGACA-- ----((((((((((.....))))))))))((.((..(((.--------((..(.....)..))..)))..)).))............-......-- ( -26.70, z-score = -0.75, R) >consensus GUACAGAUUUCCAUUAGUGGAGUGGGUUUUCCACUGCCCACGUGUGCGUGUGCUGCAUGUGUGGGCGCAAAUCGCUUGGCUCCGAAAGUUGACACG .....(((((.(...(((((((......)))))))(((((((..((((.....))))..)))))))).)))))(.((((((.....)))))))... (-26.91 = -28.50 + 1.59)

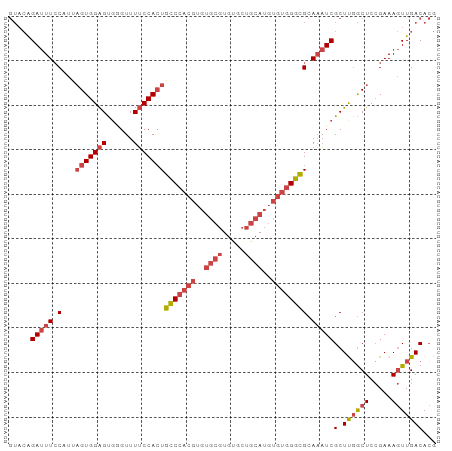
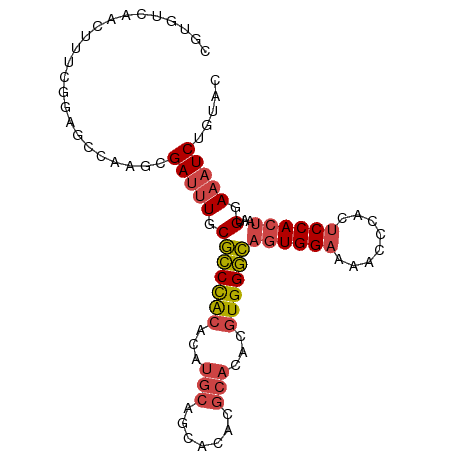
| Location | 22,118,624 – 22,118,720 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 84.10 |
| Shannon entropy | 0.30915 |
| G+C content | 0.54019 |
| Mean single sequence MFE | -29.33 |
| Consensus MFE | -21.23 |
| Energy contribution | -21.90 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.72 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.591648 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 22118624 96 - 24543557 CGUGUCAACUUUCGGAGCGAAGCGAUUUGCGCCCACACAUGCAGCACACGCACACGUGGGCAGUGGAAAACCCACUCCACUAAUGGAAAUCUGUAC .(((.((..((((...(((((....)))))((((((...(((.......)))...))))))((((((........))))))....))))..))))) ( -29.50, z-score = -1.30, R) >droSim1.chr3L 21465657 96 - 22553184 CGUGUCAACUUUCGGAGCCAAGCGAUUUGCGCCCACACAUGCAGCACACGCACACGUGGGCAGUGGAAAACCCACUCCACUAAUGGAAAUCUGUAC ............((((.(((.((.....))((((((...(((.......)))...))))))((((((........))))))..)))...))))... ( -29.40, z-score = -1.39, R) >droSec1.super_11 2048032 96 - 2888827 CGUGUCAACUUUCGGAGCCAAGCGAUUUGCGCCCACACAUGCAGCACACGCACACGUGGGCAGUGGAAAACCCACUCCACUAAUGGAAAUCUGUAC ............((((.(((.((.....))((((((...(((.......)))...))))))((((((........))))))..)))...))))... ( -29.40, z-score = -1.39, R) >droYak2.chr3L 22462000 96 - 24197627 CGUGUCAACUUUCGGAGCCAAGCGAUUUGCGCCCACACAUGCAGCACACGCACACGUGGGCAGUGGAAAACACACUCCACUAAUGGAAAUCUGUAC ............((((.(((.((.....))((((((...(((.......)))...))))))((((((........))))))..)))...))))... ( -29.40, z-score = -1.53, R) >droEre2.scaffold_4784 21789529 96 - 25762168 CGUGUCAACUUUCGGAGCCGAGCGAUUUGCGCCCACACAUGCAGCACACGCACACGUGGGCAGUGGAAAACACACUCCACUAAUGGAAAUCUGUAC .(((.((...((((....)))).(((((.(((((((...(((.......)))...))))))((((((........))))))...).)))))))))) ( -30.10, z-score = -1.51, R) >droAna3.scaffold_13337 21283716 81 - 23293914 --UGUCAA-UUUCUGAGUCGAGCGAGAUUCCUCUGCACGUGUUGCACA--------UGGAGACUGGCAAACUUGCCCCACACGGGCAAGUCC---- --(((((.-((((((((((......)))))...((((.....))))..--------.))))).))))).((((((((.....))))))))..---- ( -28.20, z-score = -1.78, R) >consensus CGUGUCAACUUUCGGAGCCAAGCGAUUUGCGCCCACACAUGCAGCACACGCACACGUGGGCAGUGGAAAACCCACUCCACUAAUGGAAAUCUGUAC .......................(((((.(((((((...(((.......)))...))))))((((((........))))))...).)))))..... (-21.23 = -21.90 + 0.67)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:43:49 2011