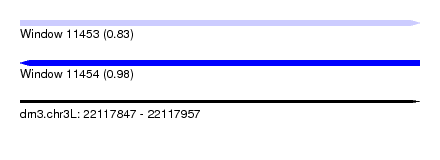
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 22,117,847 – 22,117,957 |
| Length | 110 |
| Max. P | 0.984748 |

| Location | 22,117,847 – 22,117,957 |
|---|---|
| Length | 110 |
| Sequences | 7 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 67.04 |
| Shannon entropy | 0.66166 |
| G+C content | 0.43907 |
| Mean single sequence MFE | -28.13 |
| Consensus MFE | -11.08 |
| Energy contribution | -13.66 |
| Covariance contribution | 2.58 |
| Combinations/Pair | 1.52 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.39 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.83 |
| SVM RNA-class probability | 0.830181 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
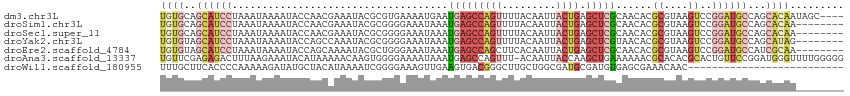
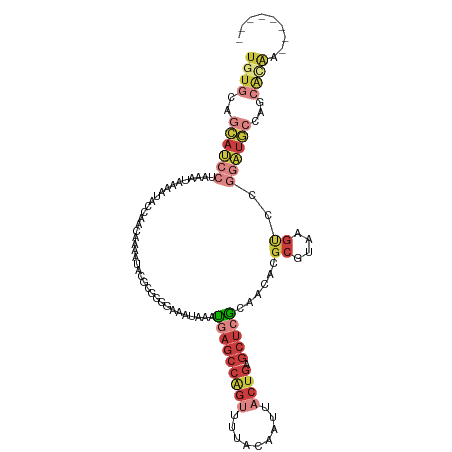
>dm3.chr3L 22117847 110 + 24543557 ----GCUAUUGUGCUGGCAUCCGGACUUACGCGUGUUGCGAGCUCAGUAAUUGUAAAACUGGCUCAUUCAUUUUCACGCGUAUUUCGUUGGUAUUUUAUUUAGGAUGCUGCACA ----.....(((((.(((((((.((..((((((((.((.((((.((((.........))))))))...))....))))))))..))..(((........))))))))))))))) ( -39.70, z-score = -4.12, R) >droSim1.chr3L 21464859 106 + 22553184 --------UUGUGCUGGCAUCCGGACUUACGCGUGUUGCGAGCUCAGUAAUUGUAAAACUGGCUCAUUUAUUUCCCCGCGUAUUUCGUUGGUAUUUUAUUUAGGAUGCUGCACA --------.(((((.((((((((((.....((.....))((((.((((.........)))))))).......)))(((((.....)).)))...........)))))))))))) ( -34.60, z-score = -2.72, R) >droSec1.super_11 2047213 106 + 2888827 --------UUGUGCUGGCAUCCGGACUUACGCGUGUUGCGAGCUCAGUAAUUGUAAAACUGGCUCAUUUAUUUCCCCGCGUAUUUCGUUGGUAUUUUAUUUAGGAUGCUGCACA --------.(((((.((((((((((.....((.....))((((.((((.........)))))))).......)))(((((.....)).)))...........)))))))))))) ( -34.60, z-score = -2.72, R) >droYak2.chr3L 22461179 106 + 24197627 --------CUAUGCUGGCAUCCGGACUUACGCGUGUUACGAGCUCAGUAAUUGUAAAACUGGCUCAUUUAUUUCCCCGCGUAUUUGGCUGGUAUUUUAUUUAGGAUGCUACACA --------...((.((((((((.(((........)))..((((.((((.........)))))))).........((.((.......)).))...........)))))))))).. ( -26.40, z-score = -0.67, R) >droEre2.scaffold_4784 21788752 106 + 25762168 --------UUGCGAUGGCAUCCGGACUUACGCGUGUUGCGAGCUCAGUAAUUGUGAAGCUGGCUCAUUUAUUUCCCAGCGUAUUUUGCUGGUAUUUUAUUUAGGAUGCUACACA --------.((.(.(((((((((((.....((.....))((((.((((.........)))))))).......)))(((((.....)))))............))))))))).)) ( -32.90, z-score = -1.83, R) >droAna3.scaffold_13337 21280812 113 + 23293914 CCCCCAAAACCCAUCCGGAACAGUGCGUGUGCGUUUUUUCAGCUUGGUAAUUGU-AAACUGGCUCAUUUAUUUUCCCCACUUGUUUUUAUGUAUUUCUUAAAGUCUCUCGAACA .............((.(((((..((.(.((((((.....(((..(((..(..((-(((........)))))..)..))).))).....))))))..).))..)).))).))... ( -10.90, z-score = 2.40, R) >droWil1.scaffold_180955 11704 88 + 2875958 --------------------------GUUGUUUCGCUCACAUCGCAUCGCCAGCAAGCCCGUCACUUCAACUUUCCCCGAUUUUAUGUAGCAUAUCUUUUUGGGGUGAAGCAAA --------------------------.(((((((((.......((.......)).....................(((((...((((...)))).....)))))))))))))). ( -17.80, z-score = -0.63, R) >consensus ________UUGUGCUGGCAUCCGGACUUACGCGUGUUGCGAGCUCAGUAAUUGUAAAACUGGCUCAUUUAUUUCCCCGCGUAUUUCGUUGGUAUUUUAUUUAGGAUGCUGCACA ..............((((((((.................((((.((((.........)))))))).........((.(((.....))).))...........)))))))).... (-11.08 = -13.66 + 2.58)

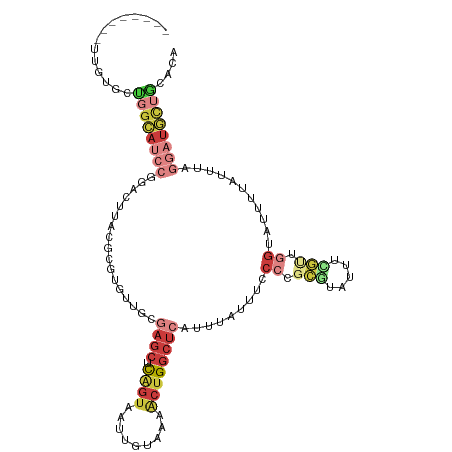
| Location | 22,117,847 – 22,117,957 |
|---|---|
| Length | 110 |
| Sequences | 7 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 67.04 |
| Shannon entropy | 0.66166 |
| G+C content | 0.43907 |
| Mean single sequence MFE | -28.34 |
| Consensus MFE | -10.01 |
| Energy contribution | -12.57 |
| Covariance contribution | 2.56 |
| Combinations/Pair | 1.43 |
| Mean z-score | -2.40 |
| Structure conservation index | 0.35 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.18 |
| SVM RNA-class probability | 0.984748 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 22117847 110 - 24543557 UGUGCAGCAUCCUAAAUAAAAUACCAACGAAAUACGCGUGAAAAUGAAUGAGCCAGUUUUACAAUUACUGAGCUCGCAACACGCGUAAGUCCGGAUGCCAGCACAAUAGC---- (((((.((((((................((..((((((((....((...((((((((.........)))).)))).)).))))))))..)).))))))..))))).....---- ( -35.83, z-score = -4.97, R) >droSim1.chr3L 21464859 106 - 22553184 UGUGCAGCAUCCUAAAUAAAAUACCAACGAAAUACGCGGGGAAAUAAAUGAGCCAGUUUUACAAUUACUGAGCUCGCAACACGCGUAAGUCCGGAUGCCAGCACAA-------- (((((.((((((...........((..((.....)).))(((.......((((((((.........)))).))))((.....)).....)))))))))..))))).-------- ( -32.40, z-score = -3.59, R) >droSec1.super_11 2047213 106 - 2888827 UGUGCAGCAUCCUAAAUAAAAUACCAACGAAAUACGCGGGGAAAUAAAUGAGCCAGUUUUACAAUUACUGAGCUCGCAACACGCGUAAGUCCGGAUGCCAGCACAA-------- (((((.((((((...........((..((.....)).))(((.......((((((((.........)))).))))((.....)).....)))))))))..))))).-------- ( -32.40, z-score = -3.59, R) >droYak2.chr3L 22461179 106 - 24197627 UGUGUAGCAUCCUAAAUAAAAUACCAGCCAAAUACGCGGGGAAAUAAAUGAGCCAGUUUUACAAUUACUGAGCUCGUAACACGCGUAAGUCCGGAUGCCAGCAUAG-------- (((((.((((((...........((.((.......))))(((.....((((((((((.........)))).))))))......(....))))))))))..))))).-------- ( -29.90, z-score = -2.84, R) >droEre2.scaffold_4784 21788752 106 - 25762168 UGUGUAGCAUCCUAAAUAAAAUACCAGCAAAAUACGCUGGGAAAUAAAUGAGCCAGCUUCACAAUUACUGAGCUCGCAACACGCGUAAGUCCGGAUGCCAUCGCAA-------- ((((..((((((............((((.......))))(((.......(((((((...........))).))))((.....)).....)))))))))...)))).-------- ( -31.10, z-score = -3.13, R) >droAna3.scaffold_13337 21280812 113 - 23293914 UGUUCGAGAGACUUUAAGAAAUACAUAAAAACAAGUGGGGAAAAUAAAUGAGCCAGUUU-ACAAUUACCAAGCUGAAAAAACGCACACGCACUGUUCCGGAUGGGUUUUGGGGG ...((....))((((((((....(((...((((.(((.(..............((((((-.........))))))............).)))))))....)))..)))))))). ( -16.77, z-score = 1.68, R) >droWil1.scaffold_180955 11704 88 - 2875958 UUUGCUUCACCCCAAAAAGAUAUGCUACAUAAAAUCGGGGAAAGUUGAAGUGACGGGCUUGCUGGCGAUGCGAUGUGAGCGAAACAAC-------------------------- .(..(((((((((.......((((...)))).....)))).....)))))..)..(..(((((.(((......))).)))))..)...-------------------------- ( -20.00, z-score = -0.37, R) >consensus UGUGCAGCAUCCUAAAUAAAAUACCAACAAAAUACGCGGGGAAAUAAAUGAGCCAGUUUUACAAUUACUGAGCUCGCAACACGCGUAAGUCCGGAUGCCAGCACAA________ ((((..((((((....................................(((((((((.........)))).)))))......((....))..))))))...))))......... (-10.01 = -12.57 + 2.56)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:43:46 2011