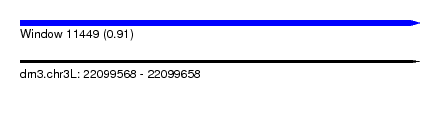
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 22,099,568 – 22,099,658 |
| Length | 90 |
| Max. P | 0.909976 |

| Location | 22,099,568 – 22,099,658 |
|---|---|
| Length | 90 |
| Sequences | 9 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 79.21 |
| Shannon entropy | 0.41029 |
| G+C content | 0.52230 |
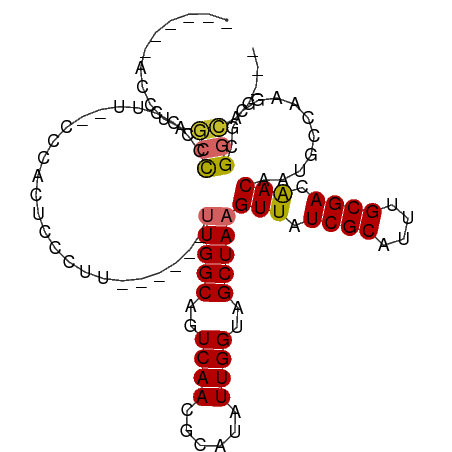
| Mean single sequence MFE | -24.18 |
| Consensus MFE | -15.89 |
| Energy contribution | -15.88 |
| Covariance contribution | -0.01 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.21 |
| SVM RNA-class probability | 0.909976 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 22099568 90 + 24543557 ------ACCUAGCCCCAAUU--CCCACUCCCUU------UUGGCAGUCAACGCAUAUUGGUAGCUAAGUUAUCGCAUUUGCGACAACAAUGCCAAACGCGGCAG--- ------...((((.(((((.--...(((.((..------..)).)))........)))))..)))).(((.((((....)))).)))..((((......)))).--- ( -22.80, z-score = -1.99, R) >droWil1.scaffold_180955 2843612 106 - 2875958 ACCGCCACCCUUCCGCUCUUUCCCCACUCCUUAGACCAGUUGGCAGUCAACGCAUAUUGGUAGCUAAGUUAUCGCAUUUGCGACAACAAUGCCA-GCGAGGAAAAAG ..........(((((.(((.............))).).(((((((.....((((.(((((((((...))))))).)).)))).......)))))-))..)))).... ( -24.32, z-score = -1.16, R) >droPer1.super_45 303423 96 - 618639 ----UGUUCUCUCCCUCUCCCACCCACUCUCUG------CUGGCAGUCAACGCAUAUUGGUAGCUAAGUUAUCGCAUUUGCGACAACAAUGCGCCGUCCGCAGUAG- ----........................(((((------(.(((.((....)).....(((.((...(((.((((....)))).)))...)))))))).)))).))- ( -21.30, z-score = -0.91, R) >dp4.chrXR_group8 6169111 96 + 9212921 ----UGUUCUCUCCCUCUCCCACCCACUCUCUG------CUGGCAGUCAACGCAUAUUGGUAGCUAAGUUAUCGCAUUUGCGACAACAAUGCGCCGUCCGCAGUAG- ----........................(((((------(.(((.((....)).....(((.((...(((.((((....)))).)))...)))))))).)))).))- ( -21.30, z-score = -0.91, R) >droAna3.scaffold_13337 21260103 98 + 23293914 ---AGAAUUCGUCUCCCCAGCGCCCACUCCCCCA-----UUGGCAGUCAACGCAUAUUGGUAGCUAAGUUAUCGCAUUUGCGACGACAAUGCCAAGCUCGGCAGAG- ---........(((.((.((((....).......-----(((((((((..((((.(((((((((...))))))).)).))))..)))..))))))))).)).))).- ( -27.00, z-score = -1.72, R) >droEre2.scaffold_4784 21771083 90 + 25762168 ------ACCUAGCCCCGAUU--CCCACUCCCCU------UUGGCAGUCAACGCGUAUUGGUAGCUAAGUUAUCGCAUUUGCGACAACAAUGCCAAGCGCGGCAG--- ------.....(((..((((--.(((.......------.))).))))...((((.((((((.....(((.((((....)))).)))..)))))))))))))..--- ( -29.80, z-score = -3.27, R) >droYak2.chr3L 22439459 90 + 24197627 ------AUCUAGCCCCAAUU--CCCACUCCCUU------UUGGCAGUCAACGCAUAUUGGUAGCUAAGUUAUCGCAUUUGCGACAACAAUGCCAAGCGCGGCAG--- ------.....(((......--...(((.((..------..)).)))....((...((((((.....(((.((((....)))).)))..))))))..)))))..--- ( -23.70, z-score = -1.84, R) >droSec1.super_11 2029151 92 + 2888827 ------ACCUAGCCCCCCUUUUCCCACUCCCUU------UUGGCAGUCAACGCAUAUUGGUAGCUAAGUUAUCGCAUUUGCGACAACAAUGCCAAGCGCGGCAG--- ------.....(((...........(((.((..------..)).)))....((...((((((.....(((.((((....)))).)))..))))))..)))))..--- ( -23.70, z-score = -1.89, R) >droSim1.chr3L 21445625 92 + 22553184 ------ACCUAGCCCCCCUUUUCCCACUCCCUU------UUGGCAGUCAACGCAUAUUGGUAGCUAAGUUAUCGCAUUUGCGACAACAAUGCCAAGCGCGGCAG--- ------.....(((...........(((.((..------..)).)))....((...((((((.....(((.((((....)))).)))..))))))..)))))..--- ( -23.70, z-score = -1.89, R) >consensus ______ACCUAGCCCCCCUU__CCCACUCCCUU______UUGGCAGUCAACGCAUAUUGGUAGCUAAGUUAUCGCAUUUGCGACAACAAUGCCAAGCGCGGCAG___ ...........(((.........................(((((..((((......))))..)))))(((.((((....)))).)))............)))..... (-15.89 = -15.88 + -0.01)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:43:42 2011