| Sequence ID | dm3.chr3L |
|---|---|
| Location | 22,064,942 – 22,065,064 |
| Length | 122 |
| Max. P | 0.777040 |

| Location | 22,064,942 – 22,065,064 |
|---|---|
| Length | 122 |
| Sequences | 5 |
| Columns | 125 |
| Reading direction | forward |
| Mean pairwise identity | 73.07 |
| Shannon entropy | 0.46922 |
| G+C content | 0.49014 |
| Mean single sequence MFE | -35.70 |
| Consensus MFE | -21.20 |
| Energy contribution | -21.45 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.09 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.530212 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
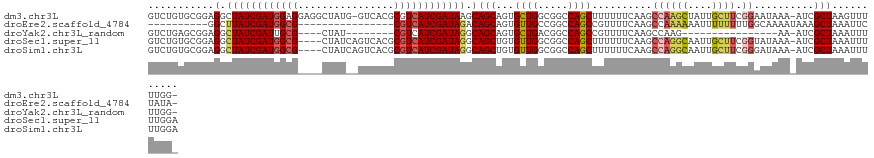
>dm3.chr3L 22064942 122 + 24543557 GUCUGUGCGGAGGCUAUCGAUGGAGGAGGCUAUG-GUCACGCGUCAUCGAUAAGCAGCAGUGCUGGCGGCCAGCUUUUUUCAAGCCAAGCUAUUGCUUCGGAAUAAA-AUCGCUAAGUUUUUGG- (((.(..(.(..((((((((((..((.(((....-))).).)..))))))).)))..).)..).)))....(((..((((....((((((....)))).))...)))-)..)))..........- ( -35.40, z-score = 0.78, R) >droEre2.scaffold_4784 21741307 98 + 25762168 ----------GGCUUAUCGAUGGCG----------------CGUCAUCGAUAGACAGCAGUGUUGCCGGCCAGCCGUUUUCAAGCCAAAAAAUUUUUUUUGGCAAAAUAAAGCUAAAUUCUAUA- ----------.(..(((((((((..----------------..)))))))))..)(((..((((..(((....))).......((((((((....))))))))..))))..)))..........- ( -28.30, z-score = -2.12, R) >droYak2.chr3L_random 217157 95 + 4797643 GUCUGAGCGGAGGCUAUCGAUUGCG----CUAU--------CGUCAUCGAUAGGCAGCAGUGCUGACGGCCAGCCGUUUUCAAGCCAAG----------------AA-AUCGCUAAAUUUUUGG- .....(((((.(.((((((((.(((----....--------))).)))))))).).((..((..(((((....)))))..)).))....----------------..-.)))))..........- ( -29.90, z-score = -0.68, R) >droSec1.super_11 1999721 120 + 2888827 GUCUGUGCGGAGGCUAUCGAUGGCG----CUAUCAGUCACGCGUCAUCGAUAGGCAGCUGUGUUGGCGGCCAGCUUUUUUCAAGCCAGGCAAUUGCUUCGGUAUAAA-AUCGCUAAAUUUUUGGA ......((((..(((((((((((((----(..........)))))))))))).))..)))).(((((((((.((((.....))))..)))...(((....)))....-..))))))......... ( -43.60, z-score = -2.18, R) >droSim1.chr3L 21411377 120 + 22553184 GUCUGUGCGGAGGCUAUCGAUGGCG----CUAUCAGUCACGCGUCAUCGAUAGGCAGCUGUGUUGGCGGCCAGCUUUUUUCAAGCCAGGCAAUUGCUUCGGGAUAAA-AUCGCUAAAUUUUUGGA ......((((..(((((((((((((----(..........)))))))))))).))..)))).(((((((((.((((.....))))..))).....(....)......-..))))))......... ( -41.30, z-score = -1.22, R) >consensus GUCUGUGCGGAGGCUAUCGAUGGCG____CUAU__GUCACGCGUCAUCGAUAGGCAGCAGUGUUGGCGGCCAGCUUUUUUCAAGCCAAGCAAUUGCUUCGGGAUAAA_AUCGCUAAAUUUUUGG_ ...........(.((((((((((((................)))))))))))).)(((...((((.....))))............((((....)))).............)))........... (-21.20 = -21.45 + 0.25)
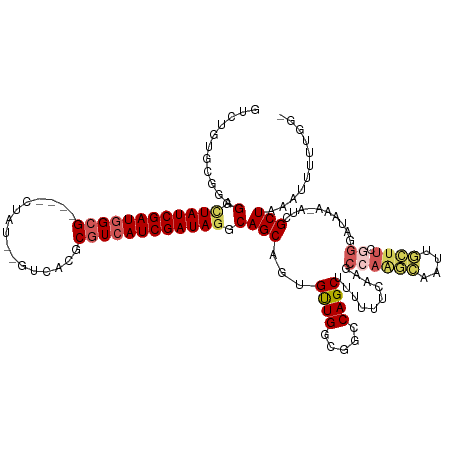

| Location | 22,064,942 – 22,065,064 |
|---|---|
| Length | 122 |
| Sequences | 5 |
| Columns | 125 |
| Reading direction | reverse |
| Mean pairwise identity | 73.07 |
| Shannon entropy | 0.46922 |
| G+C content | 0.49014 |
| Mean single sequence MFE | -29.96 |
| Consensus MFE | -16.31 |
| Energy contribution | -16.71 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.777040 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 22064942 122 - 24543557 -CCAAAAACUUAGCGAU-UUUAUUCCGAAGCAAUAGCUUGGCUUGAAAAAAGCUGGCCGCCAGCACUGCUGCUUAUCGAUGACGCGUGAC-CAUAGCCUCCUCCAUCGAUAGCCUCCGCACAGAC -...........(((.(-((((..((.((((....))))))..)))))...(((((...)))))......(((.(((((((..((.....-....))......))))))))))...)))...... ( -29.20, z-score = -0.57, R) >droEre2.scaffold_4784 21741307 98 - 25762168 -UAUAGAAUUUAGCUUUAUUUUGCCAAAAAAAAUUUUUUGGCUUGAAAACGGCUGGCCGGCAACACUGCUGUCUAUCGAUGACG----------------CGCCAUCGAUAAGCC---------- -..........((((((.((..((((((((....))))))))..)).)).))))((((((((....)))))..((((((((...----------------...)))))))).)))---------- ( -29.30, z-score = -2.86, R) >droYak2.chr3L_random 217157 95 - 4797643 -CCAAAAAUUUAGCGAU-UU----------------CUUGGCUUGAAAACGGCUGGCCGUCAGCACUGCUGCCUAUCGAUGACG--------AUAG----CGCAAUCGAUAGCCUCCGCUCAGAC -(((((((((....)))-))----------------.)))).((((...(((..(((..(((((...)))((((((((....))--------))))----.))....))..))).))).)))).. ( -25.50, z-score = -0.56, R) >droSec1.super_11 1999721 120 - 2888827 UCCAAAAAUUUAGCGAU-UUUAUACCGAAGCAAUUGCCUGGCUUGAAAAAAGCUGGCCGCCAACACAGCUGCCUAUCGAUGACGCGUGACUGAUAG----CGCCAUCGAUAGCCUCCGCACAGAC ............(((..-...........(((...(((.(((((.....)))))))).((.......)))))(((((((((.(((..........)----)).)))))))))....)))...... ( -32.90, z-score = -2.01, R) >droSim1.chr3L 21411377 120 - 22553184 UCCAAAAAUUUAGCGAU-UUUAUCCCGAAGCAAUUGCCUGGCUUGAAAAAAGCUGGCCGCCAACACAGCUGCCUAUCGAUGACGCGUGACUGAUAG----CGCCAUCGAUAGCCUCCGCACAGAC ............(((..-...........(((...(((.(((((.....)))))))).((.......)))))(((((((((.(((..........)----)).)))))))))....)))...... ( -32.90, z-score = -1.85, R) >consensus _CCAAAAAUUUAGCGAU_UUUAUCCCGAAGCAAUUGCUUGGCUUGAAAAAAGCUGGCCGCCAACACUGCUGCCUAUCGAUGACGCGUGAC__AUAG____CGCCAUCGAUAGCCUCCGCACAGAC ..........((((.......................(..((((.....))))..)..(....)...)))).(((((((((.((................)).)))))))))............. (-16.31 = -16.71 + 0.40)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:43:38 2011