
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 22,043,280 – 22,043,363 |
| Length | 83 |
| Max. P | 0.688321 |

| Location | 22,043,280 – 22,043,363 |
|---|---|
| Length | 83 |
| Sequences | 10 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 86.95 |
| Shannon entropy | 0.25700 |
| G+C content | 0.34764 |
| Mean single sequence MFE | -17.62 |
| Consensus MFE | -12.22 |
| Energy contribution | -12.97 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.69 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.688321 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
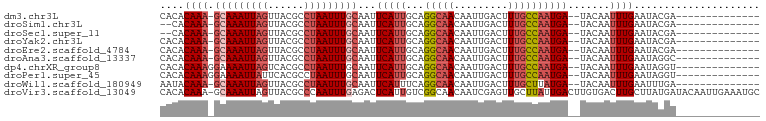
>dm3.chr3L 22043280 83 - 24543557 CACACAAA-GCAAAUUAGUUACGCCUAAUUUGCAAUUCAUUGCAGGCAACAAUUGACUUUGCCAAUGA--UACAAUUUGAAUACGA-------------- ....((((-(((((((((......)))))))))...(((((...(((((.........))))))))))--.....)))).......-------------- ( -18.30, z-score = -2.69, R) >droSim1.chr3L 21389088 81 - 22553184 --CACAAA-GCAAAUUAGUUACGCCUAAUUUGCAAUUCAUUGCAGGCAACAAUUGACUUUGCCAAUGA--UACAAUUUGAAUACGA-------------- --..((((-(((((((((......)))))))))...(((((...(((((.........))))))))))--.....)))).......-------------- ( -18.30, z-score = -2.61, R) >droSec1.super_11 1978092 81 - 2888827 --CACAAA-GCAAAUUAGUUACGCCUAAUUUGCAAUUCAUUGCAGGCAACAAUUGACUUUGCCAAUGA--UACAAUUUGAAUACGA-------------- --..((((-(((((((((......)))))))))...(((((...(((((.........))))))))))--.....)))).......-------------- ( -18.30, z-score = -2.61, R) >droYak2.chr3L 22387882 83 - 24197627 CACACAAA-GCAAAUUAGUUACGCCUAAUUUGCAAUUCAUUGCAGGCAACAAUUGACUUUGCCAAUGA--UACAAUUUGAAUACGA-------------- ....((((-(((((((((......)))))))))...(((((...(((((.........))))))))))--.....)))).......-------------- ( -18.30, z-score = -2.69, R) >droEre2.scaffold_4784 21718913 83 - 25762168 CACACAAA-GCAAAUUAGUUACGCCUAAUUUGCAAUUCAUUGCAGGCAACAAUUGACUUUGCCAAUGA--UACAAUUUGAAUACGA-------------- ....((((-(((((((((......)))))))))...(((((...(((((.........))))))))))--.....)))).......-------------- ( -18.30, z-score = -2.69, R) >droAna3.scaffold_13337 21180225 83 - 23293914 CACACAAA-GCAAAUUAGUUACGCCUAAUUUGCAAUUCAUUGCAGGCAACAAUUGACUUUGCCAAUGA--UACAAUUUGAAUAGGC-------------- ....((((-(((((((((......)))))))))...(((((...(((((.........))))))))))--.....)))).......-------------- ( -18.30, z-score = -1.98, R) >dp4.chrXR_group8 6118776 84 - 9212921 CACACAAAGGAAAAUUAGUCACGCCUAAUUUGCAAUUCAUUGCAGGCAACAAUUGACUUUGCCAAUGA--UACAAUUUGAAUAGGU-------------- ......................(((((...(((((....)))))(((((.........))))).....--...........)))))-------------- ( -14.10, z-score = -0.36, R) >droPer1.super_45 252951 84 + 618639 CACACAAAGGAAAAUUAUUCACGCCUAAUUUGCAAUUCAUUGCAGGCAACAAUUGACUUUGCCAAUGA--UACAAUUUGAAUAGGU-------------- ......................(((((...(((((....)))))(((((.........))))).....--...........)))))-------------- ( -14.10, z-score = -0.69, R) >droWil1.scaffold_180949 4649855 83 + 6375548 AAUACAAA-GCAAAUUAGUUACGCCUAAUUUGCAAUUCAUUUCAGGCAACAAUUGACUUUGCUUAUGA--UACAAUUUGAAUUUGA-------------- ....((((-(((((((((......)))))))))...((((...((((((.........))))))))))--.....)))).......-------------- ( -16.30, z-score = -1.84, R) >droVir3.scaffold_13049 21038144 99 - 25233164 CACACAAA-GCAAAUUAGUUACGCCCAAUUUGAGACUCAUUGUCGGCAACAAUCGAGUUGCUUAUUGACUUGUGACUUGCUUAUGAUACAAUUGAAAUGC ....((((-(((....(((((((..((((..(..(((((((((.....))))).))))..)..))))...)))))))))))).))............... ( -21.90, z-score = -1.31, R) >consensus CACACAAA_GCAAAUUAGUUACGCCUAAUUUGCAAUUCAUUGCAGGCAACAAUUGACUUUGCCAAUGA__UACAAUUUGAAUACGA______________ ....((((.(((((((((......)))))))))...(((((...(((((.........)))))))))).......))))..................... (-12.22 = -12.97 + 0.75)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:43:35 2011