
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 22,006,153 – 22,006,256 |
| Length | 103 |
| Max. P | 0.666015 |

| Location | 22,006,153 – 22,006,256 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 87.65 |
| Shannon entropy | 0.22486 |
| G+C content | 0.36146 |
| Mean single sequence MFE | -28.20 |
| Consensus MFE | -21.32 |
| Energy contribution | -21.18 |
| Covariance contribution | -0.13 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.76 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.605109 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
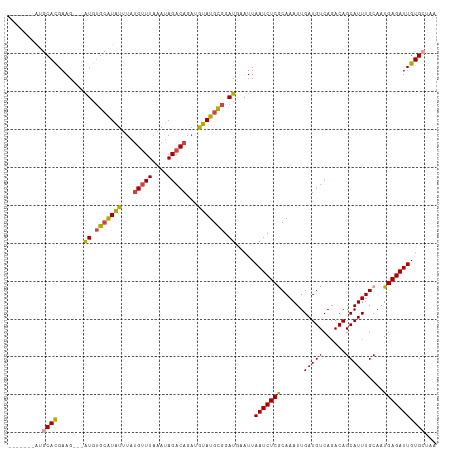
>dm3.chr3L 22006153 103 + 24543557 -------AUGCACGAAG---AUGUGCAUAUUUAUGUUUAAAUAGACAGUUGUAUGCGGAUGAAUUAAUCUCGCAAAUUGAUGUCAGACAGCAUUUGCAAUGAGAUUGUGAGAA -------...(((....---((.(((((((...(((((....)))))...))))))).)).....((((((((((((...((.....))..))))))...))))))))).... ( -25.30, z-score = -1.00, R) >droSim1.chr3L_random 933358 103 + 1049610 -------GUGCACGAAG---AUGUGCAUAUUUAUGUUUAAAUAGACAGAUGUAUGCGGAUGAAUUAAUCUCGCAAAUUGAUGUCAGACAGCAUUUGCAAUGAGAUUGUGCUAA -------..((((....---((.(((((((...(((((....)))))...))))))).)).....((((((((((((...((.....))..))))))...))))))))))... ( -28.60, z-score = -1.75, R) >droSec1.super_11 1932603 103 + 2888827 -------GUGCACGAAG---AUGUGCAUAUUUAUGUUUAAAUAGACAGAUGUAUGCGGAUGAAUUAAUCUCGCAAAUUGAUGUCAGACAGCAUUUGCAAUGAGAUUGUGCUAA -------..((((....---((.(((((((...(((((....)))))...))))))).)).....((((((((((((...((.....))..))))))...))))))))))... ( -28.60, z-score = -1.75, R) >droYak2.chr3L 22342732 103 + 24197627 -------AGGCACGAAG---AUGUGCAUAUUUAUGUUUAAAUAGACAGAUGUAUGCGGAUAAAUUAAUCUCACAAAUUGAUGUCAGACAGCAUUUGCAAUGAGAUUGUGCUAA -------.(((((....---((.(((((((...(((((....)))))...))))))).)).....(((((((......(((((......))))).....)))))))))))).. ( -29.10, z-score = -2.82, R) >droEre2.scaffold_4784 21680650 103 + 25762168 -------AUGCACGAAG---AUGUGCAUGUUUAUGUUUAAAUAGACAGAUGUAUGUGGAUGAAUUAAUCUCGCAAAUUGAUGUCACACAGCAUUUGCAAUGAGAUUGUGCUAG -------..((((((..---...(((((((((((......)))))))(((((.(((((((..((((((.......))))))))).)))))))))))))......))))))... ( -26.00, z-score = -0.61, R) >droAna3.scaffold_13337 21142544 112 + 23293914 CCGGAAGACACAUGCAGUUCGUGCGAGUGUCUCCGAUUAUAUAGACA-AUAUAUGCAGAUGAAUUAAUCUCACAAAUUGAUGUCAGACAGCAUUUGCAAUGAGAUUGUGCCAA .((((.(((((.((((.....)))).))))))))).........(((-((...((((((((......(((.(((......))).)))...)))))))).....)))))..... ( -31.60, z-score = -2.25, R) >consensus _______AUGCACGAAG___AUGUGCAUAUUUAUGUUUAAAUAGACAGAUGUAUGCGGAUGAAUUAAUCUCGCAAAUUGAUGUCAGACAGCAUUUGCAAUGAGAUUGUGCUAA .........((((.......((.(((((((...(((((....)))))...))))))).)).....(((((((......(((((......))))).....)))))))))))... (-21.32 = -21.18 + -0.13)


| Location | 22,006,153 – 22,006,256 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 87.65 |
| Shannon entropy | 0.22486 |
| G+C content | 0.36146 |
| Mean single sequence MFE | -22.03 |
| Consensus MFE | -15.87 |
| Energy contribution | -16.73 |
| Covariance contribution | 0.86 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.72 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.666015 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
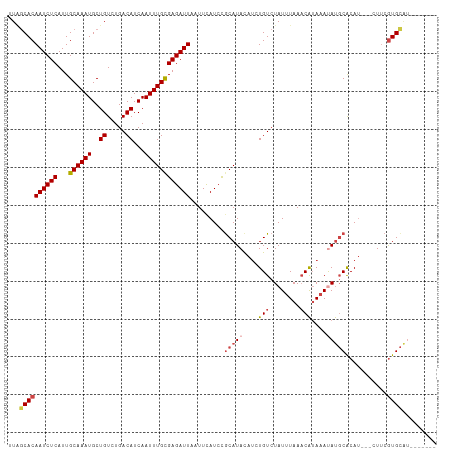
>dm3.chr3L 22006153 103 - 24543557 UUCUCACAAUCUCAUUGCAAAUGCUGUCUGACAUCAAUUUGCGAGAUUAAUUCAUCCGCAUACAACUGUCUAUUUAAACAUAAAUAUGCACAU---CUUCGUGCAU------- ....(((((((((...((((((..((.....))...)))))))))))).........(((((....(((........)))....)))))....---....)))...------- ( -17.70, z-score = -1.53, R) >droSim1.chr3L_random 933358 103 - 1049610 UUAGCACAAUCUCAUUGCAAAUGCUGUCUGACAUCAAUUUGCGAGAUUAAUUCAUCCGCAUACAUCUGUCUAUUUAAACAUAAAUAUGCACAU---CUUCGUGCAC------- ...((((((((((...((((((..((.....))...)))))))))))).........(((((....(((........)))....)))))....---....))))..------- ( -22.50, z-score = -2.53, R) >droSec1.super_11 1932603 103 - 2888827 UUAGCACAAUCUCAUUGCAAAUGCUGUCUGACAUCAAUUUGCGAGAUUAAUUCAUCCGCAUACAUCUGUCUAUUUAAACAUAAAUAUGCACAU---CUUCGUGCAC------- ...((((((((((...((((((..((.....))...)))))))))))).........(((((....(((........)))....)))))....---....))))..------- ( -22.50, z-score = -2.53, R) >droYak2.chr3L 22342732 103 - 24197627 UUAGCACAAUCUCAUUGCAAAUGCUGUCUGACAUCAAUUUGUGAGAUUAAUUUAUCCGCAUACAUCUGUCUAUUUAAACAUAAAUAUGCACAU---CUUCGUGCCU------- ...((((((((((...((((((..((.....))...)))))))))))).........(((((....(((........)))....)))))....---....))))..------- ( -20.20, z-score = -1.92, R) >droEre2.scaffold_4784 21680650 103 - 25762168 CUAGCACAAUCUCAUUGCAAAUGCUGUGUGACAUCAAUUUGCGAGAUUAAUUCAUCCACAUACAUCUGUCUAUUUAAACAUAAACAUGCACAU---CUUCGUGCAU------- ...((((((((((...((((((..((.....))...))))))))))))..................(((.(((......))).))).......---....))))..------- ( -17.30, z-score = -0.30, R) >droAna3.scaffold_13337 21142544 112 - 23293914 UUGGCACAAUCUCAUUGCAAAUGCUGUCUGACAUCAAUUUGUGAGAUUAAUUCAUCUGCAUAUAU-UGUCUAUAUAAUCGGAGACACUCGCACGAACUGCAUGUGUCUUCCGG .(((.(((((.....((((.(((..((((.(((......))).)))).....))).))))...))-))))))......(((((((((..(((.....)))..))))).)))). ( -32.00, z-score = -2.85, R) >consensus UUAGCACAAUCUCAUUGCAAAUGCUGUCUGACAUCAAUUUGCGAGAUUAAUUCAUCCGCAUACAUCUGUCUAUUUAAACAUAAAUAUGCACAU___CUUCGUGCAU_______ ...((((((((((...((((((..((.....))...)))))))))))).........(((((....(((........)))....)))))...........))))......... (-15.87 = -16.73 + 0.86)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:43:31 2011