
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 21,956,435 – 21,956,538 |
| Length | 103 |
| Max. P | 0.836845 |

| Location | 21,956,435 – 21,956,538 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 85.77 |
| Shannon entropy | 0.24828 |
| G+C content | 0.47897 |
| Mean single sequence MFE | -20.32 |
| Consensus MFE | -13.03 |
| Energy contribution | -13.23 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.512115 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
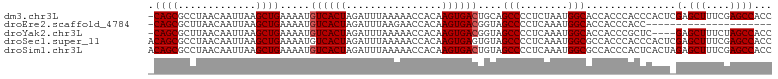
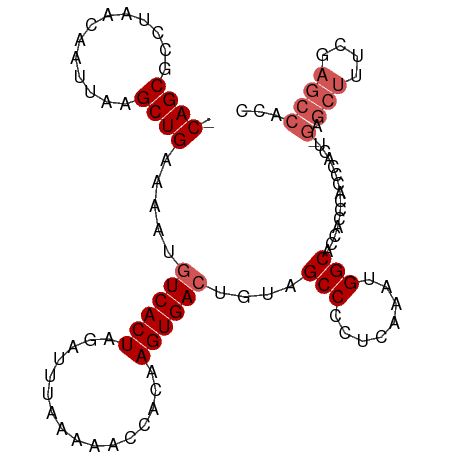
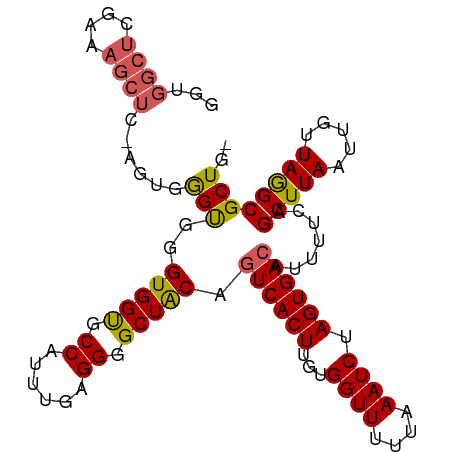
>dm3.chr3L 21956435 103 + 24543557 -CAGCGCCUAACAAUUAAGCUGAAAAUGUCACUAGAUUUAAAAACCACAAGUGACUGCAGCCCCUCUAAUGGCACCACCCACCCACUCGAGCUUUCGAGCCACC -....(((..........((((.....((((((................))))))..)))).........)))............((((......))))..... ( -20.40, z-score = -2.25, R) >droEre2.scaffold_4784 21630276 82 + 25762168 -CAGCGCUUAACAAUUAAGCUGAAAAUGUCACUAGAUUUAAGAACCACAAGUGACGGUAGCCCCUCAAAUGGCACCACCCACC--------------------- -((((.............)))).....((((((................))))))(((.(((........)))))).......--------------------- ( -18.01, z-score = -2.34, R) >droYak2.chr3L 22279831 99 + 24197627 -CAGCGCUUAACAAUUAAGCUGAAAAUGUCACUAGAUUUAAAAACCACAAGUGACGGUAGCCCCUCAAAUGGCACCACCCGCUC----GAGCUUUCUAGCCACC -.((((((((.....))))).......((((((................))))))(((.(((........))))))....))).----(.(((....))))... ( -22.39, z-score = -2.12, R) >droSec1.super_11 1881929 104 + 2888827 ACAGCGCCUAACAAUUAAGCUGAAAAUGUCACUAGAUUUAAAAACCACAAGUGAGUGUAGCCCCUCAAAUGGCGCCACCCACCCACUCGAGCUUUCGAGCCACC .((((.............))))............................(((.(((..(((........)))..))).)))...((((......))))..... ( -20.12, z-score = -1.48, R) >droSim1.chr3L 21296433 104 + 22553184 ACAGCGCCUAACAAUUAAGCUGAAAAUGUCACUAGAUUUAAAAACCACAAGUGACUGUAGCCCCUCAAAUGGCGCCACCCACUCACUAGAGCUUUCGAGCCACC ...(((((..........((((.....((((((................))))))..)))).........))))).............(.(((....))))... ( -20.70, z-score = -1.70, R) >consensus _CAGCGCCUAACAAUUAAGCUGAAAAUGUCACUAGAUUUAAAAACCACAAGUGACUGUAGCCCCUCAAAUGGCACCACCCACCCACU_GAGCUUUCGAGCCACC .((((.............)))).....((((((................))))))....(((........)))............................... (-13.03 = -13.23 + 0.20)
| Location | 21,956,435 – 21,956,538 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 85.77 |
| Shannon entropy | 0.24828 |
| G+C content | 0.47897 |
| Mean single sequence MFE | -32.28 |
| Consensus MFE | -24.78 |
| Energy contribution | -24.14 |
| Covariance contribution | -0.64 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.77 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.836845 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
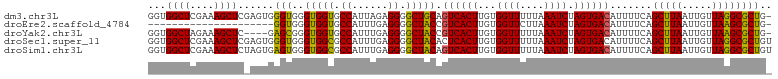
>dm3.chr3L 21956435 103 - 24543557 GGUGGCUCGAAAGCUCGAGUGGGUGGGUGGUGCCAUUAGAGGGGCUGCAGUCACUUGUGGUUUUUAAAUCUAGUGACAUUUUCAGCUUAAUUGUUAGGCGCUG- (.(.((((((....)))))).).)....((((((..(((..((((((..((((((...((((....)))).)))))).....))))))..)))...)))))).- ( -32.40, z-score = -1.24, R) >droEre2.scaffold_4784 21630276 82 - 25762168 ---------------------GGUGGGUGGUGCCAUUUGAGGGGCUACCGUCACUUGUGGUUCUUAAAUCUAGUGACAUUUUCAGCUUAAUUGUUAAGCGCUG- ---------------------(((.((((((.((......)).))))))((((((...((((....)))).)))))).......(((((.....)))))))).- ( -27.20, z-score = -2.54, R) >droYak2.chr3L 22279831 99 - 24197627 GGUGGCUAGAAAGCUC----GAGCGGGUGGUGCCAUUUGAGGGGCUACCGUCACUUGUGGUUUUUAAAUCUAGUGACAUUUUCAGCUUAAUUGUUAAGCGCUG- ...(((..((((((..----..)).((((((.((......)).))))))((((((...((((....)))).))))))..)))).(((((.....)))))))).- ( -32.80, z-score = -1.98, R) >droSec1.super_11 1881929 104 - 2888827 GGUGGCUCGAAAGCUCGAGUGGGUGGGUGGCGCCAUUUGAGGGGCUACACUCACUUGUGGUUUUUAAAUCUAGUGACAUUUUCAGCUUAAUUGUUAGGCGCUGU .(..((..((((((((((((((((..(((((.((......)).)))))))))))))).)))))))...................(((((.....)))))))..) ( -35.20, z-score = -2.02, R) >droSim1.chr3L 21296433 104 - 22553184 GGUGGCUCGAAAGCUCUAGUGAGUGGGUGGCGCCAUUUGAGGGGCUACAGUCACUUGUGGUUUUUAAAUCUAGUGACAUUUUCAGCUUAAUUGUUAGGCGCUGU .(..((..((((((((....))))..(((((.((......)).))))).((((((...((((....)))).))))))..)))).(((((.....)))))))..) ( -33.80, z-score = -1.69, R) >consensus GGUGGCUCGAAAGCUC_AGUGGGUGGGUGGUGCCAUUUGAGGGGCUACAGUCACUUGUGGUUUUUAAAUCUAGUGACAUUUUCAGCUUAAUUGUUAGGCGCUG_ ...((((....))))......(((..(((((.((......)).))))).((((((...((((....)))).)))))).......(((((.....)))))))).. (-24.78 = -24.14 + -0.64)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:43:22 2011