| Sequence ID | dm3.chr3L |
|---|---|
| Location | 21,922,376 – 21,922,468 |
| Length | 92 |
| Max. P | 0.993417 |

| Location | 21,922,376 – 21,922,468 |
|---|---|
| Length | 92 |
| Sequences | 8 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 85.70 |
| Shannon entropy | 0.27653 |
| G+C content | 0.40742 |
| Mean single sequence MFE | -24.23 |
| Consensus MFE | -15.36 |
| Energy contribution | -15.52 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.09 |
| Mean z-score | -3.36 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.61 |
| SVM RNA-class probability | 0.993417 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
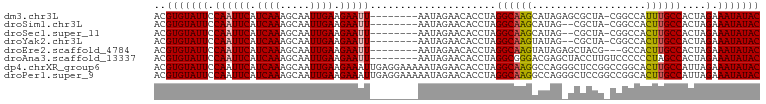
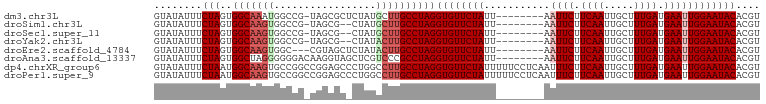
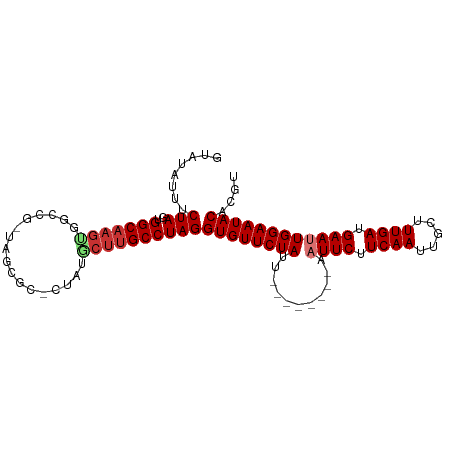
>dm3.chr3L 21922376 92 + 24543557 ACGUGUAUUCCAAUUCAUCAAAGCAAUUGAAGAAUU--------AAUAGAACACCUAGGCAAGCAUAGAGCGCUA-CGGCCAUUUGCCACUAGAAAUAUAC ..(((((((.((((((.((((.....)))).)))))--------.............(((((((.....))((..-..))...)))))....).))))))) ( -21.10, z-score = -2.85, R) >droSim1.chr3L 21263063 90 + 22553184 ACGUGUAUUCCAAUUCAUCAAAGCAAUUGAAGAAUU--------AAUAGAACACCUAGGCAAGCAUAG--CGCUA-CGGCCACUUGCCACUAGAAAUAUAC ..(((((((.((((((.((((.....)))).)))))--------.............((((((....(--.((..-..))).))))))....).))))))) ( -21.20, z-score = -3.07, R) >droSec1.super_11 1849507 90 + 2888827 ACGUGUAUUCCAAUUCAUCAAAGCAAUUGAAGAAUU--------AAUAGAACACCUAGGCAAGCAUAG--CGCUA-CGGCCACUUGCCACUAGAAAUAUAC ..(((((((.((((((.((((.....)))).)))))--------.............((((((....(--.((..-..))).))))))....).))))))) ( -21.20, z-score = -3.07, R) >droYak2.chr3L 22239769 90 + 24197627 ACGUGUAUUCCAAUUCAUCAAAGCAAUUGAAGAAUU--------AAUAGAACACCUAGGCAAGUAUAG--CGCUA-CGGCCACUUGCCACUAGAAAUAUAC ..(((((((.((((((.((((.....)))).)))))--------.............(((((((...(--.((..-..))))))))))....).))))))) ( -23.00, z-score = -3.73, R) >droEre2.scaffold_4784 21598191 90 + 25762168 ACGUGUAUUCCAAUUCAUCAAAGCAAUUGAAGAAUU--------AAUAGAACACCUAGGCAAGUAUAGAGCUACG---GCCACUUGCCACUAGAAAUAUAC ..(((((((.((((((.((((.....)))).)))))--------.............(((((((...(.((....---))))))))))....).))))))) ( -23.00, z-score = -4.02, R) >droAna3.scaffold_13337 21051123 93 + 23293914 ACGUGUAUUCCAAUUCAUCAAAGCAAUUGAAGAAUU--------AAUAGAACACCUAGGCGGGACGAGCUACCUUGUCCCCCCUAGCCACUAGAAAUAUAC ..(((((((.((((((.((((.....)))).)))))--------..........(((((.((((((((....)))))))).)))))......).))))))) ( -29.30, z-score = -5.46, R) >dp4.chrXR_group6 8136200 101 - 13314419 ACGUGUAUUCCAAUUCAUCAAAGCAAUUGAAGAAAUUGAGGAAAAAUAGAACACCUAGGCAAGGCCAGGGCUCCGGCCGGCACUUGCCAUUAGAAAUAUAC ..(((((((.((((((.((((.....)))).).)))))...................(((((((((..(((....)))))).))))))......))))))) ( -27.50, z-score = -2.35, R) >droPer1.super_9 149483 101 - 3637205 ACGUGUAUUCCAAUUCAUCAAAGCAAUUGAAGAAAUUGAGGAAAAAUAGAACACCUAGGCAAGGCCAGGGCUCCGGCCGGCACUUGCCAUUAGAAAUAUAC ..(((((((.((((((.((((.....)))).).)))))...................(((((((((..(((....)))))).))))))......))))))) ( -27.50, z-score = -2.35, R) >consensus ACGUGUAUUCCAAUUCAUCAAAGCAAUUGAAGAAUU________AAUAGAACACCUAGGCAAGCAUAG_GCGCUA_CGGCCACUUGCCACUAGAAAUAUAC ..(((((((.((((((.((((.....)))).))))).....................((((((...................))))))....).))))))) (-15.36 = -15.52 + 0.16)
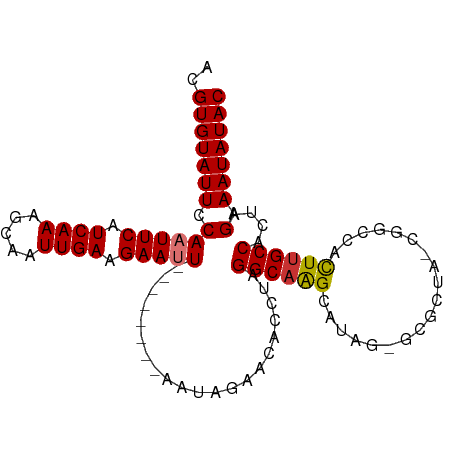
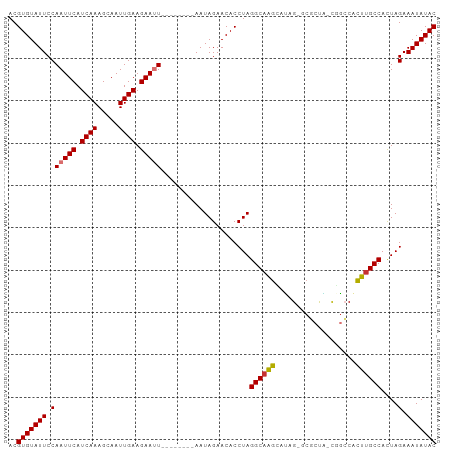
| Location | 21,922,376 – 21,922,468 |
|---|---|
| Length | 92 |
| Sequences | 8 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 85.70 |
| Shannon entropy | 0.27653 |
| G+C content | 0.40742 |
| Mean single sequence MFE | -27.33 |
| Consensus MFE | -17.04 |
| Energy contribution | -17.67 |
| Covariance contribution | 0.63 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.860662 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 21922376 92 - 24543557 GUAUAUUUCUAGUGGCAAAUGGCCG-UAGCGCUCUAUGCUUGCCUAGGUGUUCUAUU--------AAUUCUUCAAUUGCUUUGAUGAAUUGGAAUACACGU ........((((.(((.....)))(-(((.((.....))))))))))((((((((..--------.((((.((((.....)))).)))))))))))).... ( -23.40, z-score = -1.56, R) >droSim1.chr3L 21263063 90 - 22553184 GUAUAUUUCUAGUGGCAAGUGGCCG-UAGCG--CUAUGCUUGCCUAGGUGUUCUAUU--------AAUUCUUCAAUUGCUUUGAUGAAUUGGAAUACACGU ........(((..((((((((((..-....)--))..))))))))))((((((((..--------.((((.((((.....)))).)))))))))))).... ( -25.70, z-score = -2.08, R) >droSec1.super_11 1849507 90 - 2888827 GUAUAUUUCUAGUGGCAAGUGGCCG-UAGCG--CUAUGCUUGCCUAGGUGUUCUAUU--------AAUUCUUCAAUUGCUUUGAUGAAUUGGAAUACACGU ........(((..((((((((((..-....)--))..))))))))))((((((((..--------.((((.((((.....)))).)))))))))))).... ( -25.70, z-score = -2.08, R) >droYak2.chr3L 22239769 90 - 24197627 GUAUAUUUCUAGUGGCAAGUGGCCG-UAGCG--CUAUACUUGCCUAGGUGUUCUAUU--------AAUUCUUCAAUUGCUUUGAUGAAUUGGAAUACACGU ........(((..((((((((((..-....)--))..))))))))))((((((((..--------.((((.((((.....)))).)))))))))))).... ( -25.40, z-score = -2.29, R) >droEre2.scaffold_4784 21598191 90 - 25762168 GUAUAUUUCUAGUGGCAAGUGGC---CGUAGCUCUAUACUUGCCUAGGUGUUCUAUU--------AAUUCUUCAAUUGCUUUGAUGAAUUGGAAUACACGU ........(((..((((((((((---....)))....))))))))))((((((((..--------.((((.((((.....)))).)))))))))))).... ( -24.50, z-score = -2.41, R) >droAna3.scaffold_13337 21051123 93 - 23293914 GUAUAUUUCUAGUGGCUAGGGGGGACAAGGUAGCUCGUCCCGCCUAGGUGUUCUAUU--------AAUUCUUCAAUUGCUUUGAUGAAUUGGAAUACACGU ...............(((((.(((((.((....)).))))).)))))((((((((..--------.((((.((((.....)))).)))))))))))).... ( -32.70, z-score = -3.52, R) >dp4.chrXR_group6 8136200 101 + 13314419 GUAUAUUUCUAAUGGCAAGUGCCGGCCGGAGCCCUGGCCUUGCCUAGGUGUUCUAUUUUUCCUCAAUUUCUUCAAUUGCUUUGAUGAAUUGGAAUACACGU ........(((..((((((.((((((....)))..))))))))))))((((((((...(((.((((..............)))).))).)))))))).... ( -30.64, z-score = -2.59, R) >droPer1.super_9 149483 101 + 3637205 GUAUAUUUCUAAUGGCAAGUGCCGGCCGGAGCCCUGGCCUUGCCUAGGUGUUCUAUUUUUCCUCAAUUUCUUCAAUUGCUUUGAUGAAUUGGAAUACACGU ........(((..((((((.((((((....)))..))))))))))))((((((((...(((.((((..............)))).))).)))))))).... ( -30.64, z-score = -2.59, R) >consensus GUAUAUUUCUAGUGGCAAGUGGCCG_UAGCGC_CUAUGCUUGCCUAGGUGUUCUAUU________AAUUCUUCAAUUGCUUUGAUGAAUUGGAAUACACGU ........(((..(((((((.................))))))))))((((((((...........((((.((((.....)))).)))))))))))).... (-17.04 = -17.67 + 0.63)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:43:17 2011