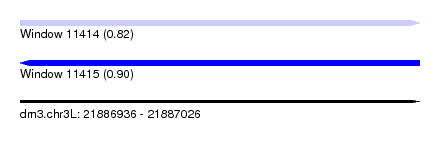
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 21,886,936 – 21,887,026 |
| Length | 90 |
| Max. P | 0.902725 |

| Location | 21,886,936 – 21,887,026 |
|---|---|
| Length | 90 |
| Sequences | 8 |
| Columns | 90 |
| Reading direction | forward |
| Mean pairwise identity | 86.50 |
| Shannon entropy | 0.27727 |
| G+C content | 0.44617 |
| Mean single sequence MFE | -18.82 |
| Consensus MFE | -13.17 |
| Energy contribution | -13.76 |
| Covariance contribution | 0.59 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.70 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.78 |
| SVM RNA-class probability | 0.816077 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
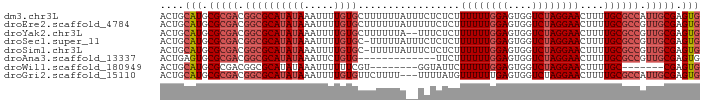
>dm3.chr3L 21886936 90 + 24543557 CACUCGCAAUGGCGCAAAAGUUCCUAGACCACUCCAAAAAAGAGAGAAAUAAAAAAGCACAAAAUUUAUAUGCGCCGUCGCGCAUGCAGU ((..(((.((((((((...(((....)))..((((......).)))........................)))))))).)))..)).... ( -19.70, z-score = -1.76, R) >droEre2.scaffold_4784 21562320 90 + 25762168 CACUCGCAACGGCGCAAAAGUUCCUAGACCACUCCAAAAAAGAGAAAAAUAAAAAAGCACAAAAUUUAUAUGCGCCGUCGCGCAUGCAGU ((..(((.((((((((...(((....)))..(((.......)))..........................)))))))).)))..)).... ( -21.00, z-score = -2.62, R) >droYak2.chr3L 22186919 88 + 24197627 CACUCGCAACGGCGCAAAAGUUCCUAGACCACUCCAAAAAAGAGAAA--UAAAAAAGCACAAAAUUUAUAUGCGCCGUCGCGCAUGCAGU ((..(((.((((((((...(((....)))..(((.......)))...--.....................)))))))).)))..)).... ( -21.00, z-score = -2.57, R) >droSec1.super_11 1814315 89 + 2888827 CACUCGCAACGGCGCAAAAGUUCCUAGACCACUCCAAAAAAGAGAGAAAUAAAAA-GCACAAAAUUUAUAUGCGCCGUCGCGCAUGCAGU ((..(((.((((((((...(((....)))..((((......).))).........-..............)))))))).)))..)).... ( -21.50, z-score = -2.45, R) >droSim1.chr3L 21225508 89 + 22553184 CACUCGCAACGGCGCAAAAGUUCCUAGACCACUCCAAAAAAGAGAGAAAUAAAAA-GCACAAAAUUUAUAUGCGCCGUCGCGCAUGCAGU ((..(((.((((((((...(((....)))..((((......).))).........-..............)))))))).)))..)).... ( -21.50, z-score = -2.45, R) >droAna3.scaffold_13337 21018803 77 + 23293914 CACUCGCAACGGCGCAAAAGUUCCUAGACCACUCCAAAAAAGAA-------------CACAGAAUUUAUAUGCGCCGUCGCGCACUCAGU ....(((.((((((((.((((((.....................-------------....))))))...)))))))).)))........ ( -20.01, z-score = -3.12, R) >droWil1.scaffold_180949 4417909 75 - 6375548 CACUCG-------GCAAAAGUUCCUAGACCACUCCAAAAAGAAUACC--------ACGAAAAAAUUUAUAUGCGCCGUCGCGCAUGCAGU ..((.(-------(........)).))....................--------.............(((((((....))))))).... ( -9.80, z-score = 0.29, R) >droGri2.scaffold_15110 1692672 87 - 24565398 CACUCGCAAUGGCGCAAAAGUUCCUAGACCACUCAAAAAACAUAAAA---AAAAGAACACAAAAUUUAUAUGCGCCGUCGCGCAUGCAGU ((..(((.((((((((...(((....)))............(((((.---..............))))).)))))))).)))..)).... ( -16.06, z-score = -1.31, R) >consensus CACUCGCAACGGCGCAAAAGUUCCUAGACCACUCCAAAAAAGAGAAA__UAAAAAAGCACAAAAUUUAUAUGCGCCGUCGCGCAUGCAGU ....(((.((((((((.(((((........................................)))))...)))))))).)))........ (-13.17 = -13.76 + 0.59)


| Location | 21,886,936 – 21,887,026 |
|---|---|
| Length | 90 |
| Sequences | 8 |
| Columns | 90 |
| Reading direction | reverse |
| Mean pairwise identity | 86.50 |
| Shannon entropy | 0.27727 |
| G+C content | 0.44617 |
| Mean single sequence MFE | -26.96 |
| Consensus MFE | -17.55 |
| Energy contribution | -18.69 |
| Covariance contribution | 1.14 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.16 |
| SVM RNA-class probability | 0.902725 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
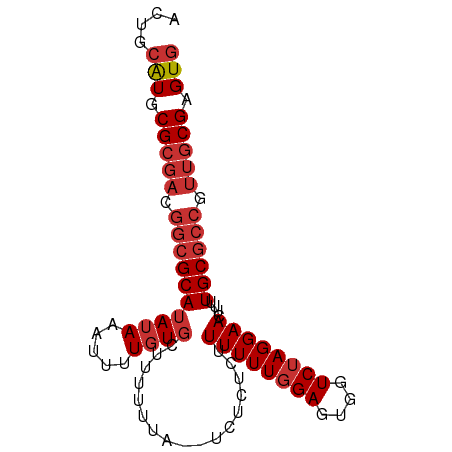
>dm3.chr3L 21886936 90 - 24543557 ACUGCAUGCGCGACGGCGCAUAUAAAUUUUGUGCUUUUUUAUUUCUCUCUUUUUUGGAGUGGUCUAGGAACUUUUGCGCCAUUGCGAGUG ((((((.((((((.((((((.........))))))((((((..((.((((.....)))).))..))))))...))))))...))).))). ( -26.20, z-score = -2.05, R) >droEre2.scaffold_4784 21562320 90 - 25762168 ACUGCAUGCGCGACGGCGCAUAUAAAUUUUGUGCUUUUUUAUUUUUCUCUUUUUUGGAGUGGUCUAGGAACUUUUGCGCCGUUGCGAGUG ....(((.((((((((((((......(((((.(((...........((((.....)))).))).))))).....)))))))))))).))) ( -29.60, z-score = -3.13, R) >droYak2.chr3L 22186919 88 - 24197627 ACUGCAUGCGCGACGGCGCAUAUAAAUUUUGUGCUUUUUUA--UUUCUCUUUUUUGGAGUGGUCUAGGAACUUUUGCGCCGUUGCGAGUG ....(((.((((((((((((......(((((.(((......--...((((.....)))).))).))))).....)))))))))))).))) ( -29.80, z-score = -3.14, R) >droSec1.super_11 1814315 89 - 2888827 ACUGCAUGCGCGACGGCGCAUAUAAAUUUUGUGC-UUUUUAUUUCUCUCUUUUUUGGAGUGGUCUAGGAACUUUUGCGCCGUUGCGAGUG ....(((.((((((((((((((((.....)))).-((((((..((.((((.....)))).))..))))))....)))))))))))).))) ( -31.30, z-score = -3.57, R) >droSim1.chr3L 21225508 89 - 22553184 ACUGCAUGCGCGACGGCGCAUAUAAAUUUUGUGC-UUUUUAUUUCUCUCUUUUUUGGAGUGGUCUAGGAACUUUUGCGCCGUUGCGAGUG ....(((.((((((((((((((((.....)))).-((((((..((.((((.....)))).))..))))))....)))))))))))).))) ( -31.30, z-score = -3.57, R) >droAna3.scaffold_13337 21018803 77 - 23293914 ACUGAGUGCGCGACGGCGCAUAUAAAUUCUGUG-------------UUCUUUUUUGGAGUGGUCUAGGAACUUUUGCGCCGUUGCGAGUG ........((((((((((((((((.....))))-------------)...((((((((....)))))))).....))))))))))).... ( -27.20, z-score = -2.39, R) >droWil1.scaffold_180949 4417909 75 + 6375548 ACUGCAUGCGCGACGGCGCAUAUAAAUUUUUUCGU--------GGUAUUCUUUUUGGAGUGGUCUAGGAACUUUUGC-------CGAGUG (((..((((((....)))))).............(--------((((...((((((((....))))))))....)))-------))))). ( -18.10, z-score = -0.34, R) >droGri2.scaffold_15110 1692672 87 + 24565398 ACUGCAUGCGCGACGGCGCAUAUAAAUUUUGUGUUCUUUU---UUUUAUGUUUUUUGAGUGGUCUAGGAACUUUUGCGCCAUUGCGAGUG ....(((.(((((.((((((............((((((..---..((((.........))))...))))))...)))))).))))).))) ( -22.16, z-score = -0.95, R) >consensus ACUGCAUGCGCGACGGCGCAUAUAAAUUUUGUGCUUUUUUA__UCUCUCUUUUUUGGAGUGGUCUAGGAACUUUUGCGCCGUUGCGAGUG ....(((.(((((.((((((((((.....)))).................((((((((....))))))))....)))))).))))).))) (-17.55 = -18.69 + 1.14)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:43:13 2011