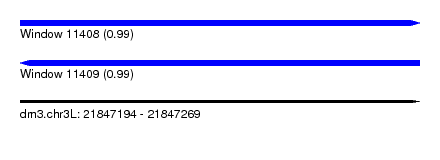
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 21,847,194 – 21,847,269 |
| Length | 75 |
| Max. P | 0.992330 |

| Location | 21,847,194 – 21,847,269 |
|---|---|
| Length | 75 |
| Sequences | 12 |
| Columns | 76 |
| Reading direction | forward |
| Mean pairwise identity | 85.41 |
| Shannon entropy | 0.34362 |
| G+C content | 0.59386 |
| Mean single sequence MFE | -27.64 |
| Consensus MFE | -26.66 |
| Energy contribution | -26.16 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.32 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.96 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.53 |
| SVM RNA-class probability | 0.992330 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
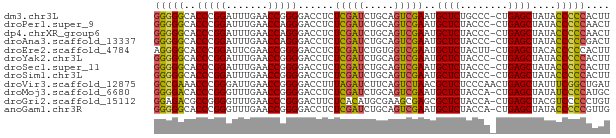
>dm3.chr3L 21847194 75 + 24543557 GGGGGCACCCGGAUUUGAACCGGGGACCUCUCGAUCUGCAGUCGAAUGCUCUGCCC-CUGAGCUAUACCCCCACUU (((((..(((((.......)))))......(((((.....)))))..((((.....-..))))....))))).... ( -28.90, z-score = -2.15, R) >droPer1.super_9 217929 75 - 3637205 GGGGGCACCCGGAUUUGAACCAGGGACCUCUCGAUCUGCAGUCGAAUGCUCUACCC-CUGAGCUAUACCCCCAACU (((((..(((((.......)).))).....(((((.....)))))..((((.....-..))))....))))).... ( -24.90, z-score = -1.61, R) >dp4.chrXR_group6 8204334 75 - 13314419 GGGGGCACCCGGAUUUGAACCAGGGACCUCUCGAUCUGCAGUCGAAUGCUCUACCC-CUGAGCUAUACCCCCAACU (((((..(((((.......)).))).....(((((.....)))))..((((.....-..))))....))))).... ( -24.90, z-score = -1.61, R) >droAna3.scaffold_13337 20974532 75 + 23293914 GGGGGCACCCGGAUUUGAACCAGGGACCUCUCGAUCUGCAGUCGAAUGCUCUACCC-CUGAGCUAUACCCCCGACU (((((..(((((.......)).))).....(((((.....)))))..((((.....-..))))....))))).... ( -24.90, z-score = -1.37, R) >droEre2.scaffold_4784 21522304 75 + 25762168 AGGGGCACCCGGAUUCGAACCGGGGACCUCUCGAUCUGUGGUCGAAUGCUCUACUU-CUGAGCUACACCCCCACUU .((((..(((((.......)))))((((.(.......).))))....((((.....-..)))).....)))).... ( -28.00, z-score = -2.12, R) >droYak2.chr3L 22114109 75 + 24197627 GGGGGCACCCGGAUUUGAACCGGGGACCUCUCGAUCUGCAGUCGAAUGCUCUACCC-CUGAGCUAUACCCCCACUU (((((..(((((.......)))))......(((((.....)))))..((((.....-..))))....))))).... ( -28.90, z-score = -2.58, R) >droSec1.super_11 1768640 75 + 2888827 GGGGGCACCCGGAUUUGAACCGGGGACCUCUCGAUCUGCAGUCGAAUGCUCUACCC-CUGAGCUAUACCCCCACUU (((((..(((((.......)))))......(((((.....)))))..((((.....-..))))....))))).... ( -28.90, z-score = -2.58, R) >droSim1.chr3L 21185264 75 + 22553184 GGGGGCACCCGGAUUUGAACCGGGGACCUCUCGAUCUGCAGUCGAAUGCUCUACCC-CUGAGCUAUACCCCCACUU (((((..(((((.......)))))......(((((.....)))))..((((.....-..))))....))))).... ( -28.90, z-score = -2.58, R) >droVir3.scaffold_12875 9400588 76 - 20611582 GCCGAAACCCGGGAUUGAACCGGGGACCUUUAGAUCUUCAGUCUAACGCUCUCCCAACUGAGCUAUUUCGGCUGAU ((((((((((((.......))))).....((((((.....)))))).((((........))))..))))))).... ( -29.10, z-score = -3.77, R) >droMoj3.scaffold_6680 996245 75 + 24764193 GGGGACACCCGGGUUUGAACCGGGGACCUCUCGAUCUGCAGUCGAAUGCUCUACCA-CUGAGCUAUAUCCCCAUGC (((((..(((((.......)))))......(((((.....)))))..((((.....-..))))....))))).... ( -28.70, z-score = -2.38, R) >droGri2.scaffold_15112 4494647 75 + 5172618 GGAGACGCCGGGGUUUGAACCCGGGACUUCUCACAUGCGAAGCGAGCGCUCUACCA-CUGAGCUACGUCCCCCUGU ((.(((((((((.......)))))(.((((.(....).)))))....((((.....-..))))..)))).)).... ( -25.40, z-score = -0.73, R) >anoGam1.chr3R 13657353 75 + 53272125 GGGGGCACCCGGGUUUGAACCGGGGACCUCUCGAUCUGCAGUCGAAUGCUCUACCA-CUGAGCUAUACCCCCGUUG (((((..(((((.......)))))......(((((.....)))))..((((.....-..))))....))))).... ( -30.20, z-score = -2.11, R) >consensus GGGGGCACCCGGAUUUGAACCGGGGACCUCUCGAUCUGCAGUCGAAUGCUCUACCC_CUGAGCUAUACCCCCACUU (((((..(((((.......)))))......(((((.....)))))..((((........))))....))))).... (-26.66 = -26.16 + -0.50)
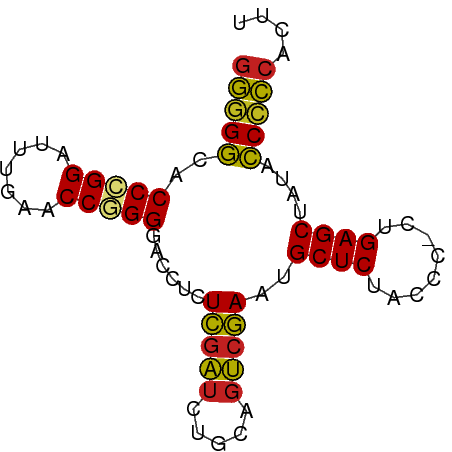
| Location | 21,847,194 – 21,847,269 |
|---|---|
| Length | 75 |
| Sequences | 12 |
| Columns | 76 |
| Reading direction | reverse |
| Mean pairwise identity | 85.41 |
| Shannon entropy | 0.34362 |
| G+C content | 0.59386 |
| Mean single sequence MFE | -29.55 |
| Consensus MFE | -30.33 |
| Energy contribution | -28.85 |
| Covariance contribution | -1.48 |
| Combinations/Pair | 1.60 |
| Mean z-score | -1.70 |
| Structure conservation index | 1.03 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.26 |
| SVM RNA-class probability | 0.986899 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 21847194 75 - 24543557 AAGUGGGGGUAUAGCUCAG-GGGCAGAGCAUUCGACUGCAGAUCGAGAGGUCCCCGGUUCAAAUCCGGGUGCCCCC ....(((((((..(((...-.)))...(((......))).((((....))))(((((.......)))))))))))) ( -31.10, z-score = -1.76, R) >droPer1.super_9 217929 75 + 3637205 AGUUGGGGGUAUAGCUCAG-GGGUAGAGCAUUCGACUGCAGAUCGAGAGGUCCCUGGUUCAAAUCCGGGUGCCCCC ....(((((((..((((..-.....))))...........((((....))))(((((.......)))))))))))) ( -28.90, z-score = -1.27, R) >dp4.chrXR_group6 8204334 75 + 13314419 AGUUGGGGGUAUAGCUCAG-GGGUAGAGCAUUCGACUGCAGAUCGAGAGGUCCCUGGUUCAAAUCCGGGUGCCCCC ....(((((((..((((..-.....))))...........((((....))))(((((.......)))))))))))) ( -28.90, z-score = -1.27, R) >droAna3.scaffold_13337 20974532 75 - 23293914 AGUCGGGGGUAUAGCUCAG-GGGUAGAGCAUUCGACUGCAGAUCGAGAGGUCCCUGGUUCAAAUCCGGGUGCCCCC ....(((((((..((((..-.....))))...........((((....))))(((((.......)))))))))))) ( -29.20, z-score = -1.23, R) >droEre2.scaffold_4784 21522304 75 - 25762168 AAGUGGGGGUGUAGCUCAG-AAGUAGAGCAUUCGACCACAGAUCGAGAGGUCCCCGGUUCGAAUCCGGGUGCCCCU ....(((((..(.((((..-.....))))((((((((...((((....))))...)).))))))....)..))))) ( -29.30, z-score = -2.06, R) >droYak2.chr3L 22114109 75 - 24197627 AAGUGGGGGUAUAGCUCAG-GGGUAGAGCAUUCGACUGCAGAUCGAGAGGUCCCCGGUUCAAAUCCGGGUGCCCCC ....(((((((..((((..-.....))))...........((((....))))(((((.......)))))))))))) ( -31.10, z-score = -2.04, R) >droSec1.super_11 1768640 75 - 2888827 AAGUGGGGGUAUAGCUCAG-GGGUAGAGCAUUCGACUGCAGAUCGAGAGGUCCCCGGUUCAAAUCCGGGUGCCCCC ....(((((((..((((..-.....))))...........((((....))))(((((.......)))))))))))) ( -31.10, z-score = -2.04, R) >droSim1.chr3L 21185264 75 - 22553184 AAGUGGGGGUAUAGCUCAG-GGGUAGAGCAUUCGACUGCAGAUCGAGAGGUCCCCGGUUCAAAUCCGGGUGCCCCC ....(((((((..((((..-.....))))...........((((....))))(((((.......)))))))))))) ( -31.10, z-score = -2.04, R) >droVir3.scaffold_12875 9400588 76 + 20611582 AUCAGCCGAAAUAGCUCAGUUGGGAGAGCGUUAGACUGAAGAUCUAAAGGUCCCCGGUUCAAUCCCGGGUUUCGGC ....(((((((..((((........))))...........((((....))))(((((.......)))))))))))) ( -25.20, z-score = -1.57, R) >droMoj3.scaffold_6680 996245 75 - 24764193 GCAUGGGGAUAUAGCUCAG-UGGUAGAGCAUUCGACUGCAGAUCGAGAGGUCCCCGGUUCAAACCCGGGUGUCCCC ....(((((((..((((..-.....))))...........((((....))))(((((.......)))))))))))) ( -28.60, z-score = -1.83, R) >droGri2.scaffold_15112 4494647 75 - 5172618 ACAGGGGGACGUAGCUCAG-UGGUAGAGCGCUCGCUUCGCAUGUGAGAAGUCCCGGGUUCAAACCCCGGCGUCUCC ....(((((((...((((.-..(..((((....))))..)...)))).....(((((.......)))))))))))) ( -29.20, z-score = -1.17, R) >anoGam1.chr3R 13657353 75 - 53272125 CAACGGGGGUAUAGCUCAG-UGGUAGAGCAUUCGACUGCAGAUCGAGAGGUCCCCGGUUCAAACCCGGGUGCCCCC ....(((((((..((((..-.....))))...........((((....))))(((((.......)))))))))))) ( -30.90, z-score = -2.19, R) >consensus AAGUGGGGGUAUAGCUCAG_GGGUAGAGCAUUCGACUGCAGAUCGAGAGGUCCCCGGUUCAAAUCCGGGUGCCCCC ....(((((((..((((........))))...........((((....))))(((((.......)))))))))))) (-30.33 = -28.85 + -1.48)

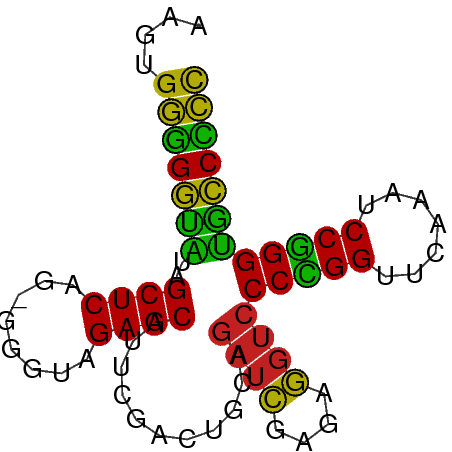
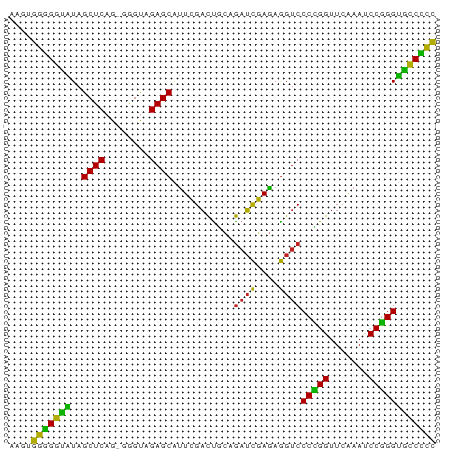
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:43:08 2011