| Sequence ID | dm3.chr3L |
|---|---|
| Location | 21,783,005 – 21,783,103 |
| Length | 98 |
| Max. P | 0.962107 |

| Location | 21,783,005 – 21,783,103 |
|---|---|
| Length | 98 |
| Sequences | 11 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 86.70 |
| Shannon entropy | 0.28262 |
| G+C content | 0.44323 |
| Mean single sequence MFE | -23.13 |
| Consensus MFE | -17.20 |
| Energy contribution | -17.05 |
| Covariance contribution | -0.15 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.74 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.70 |
| SVM RNA-class probability | 0.962107 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
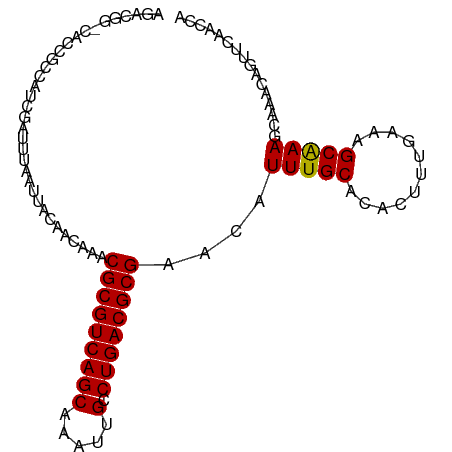
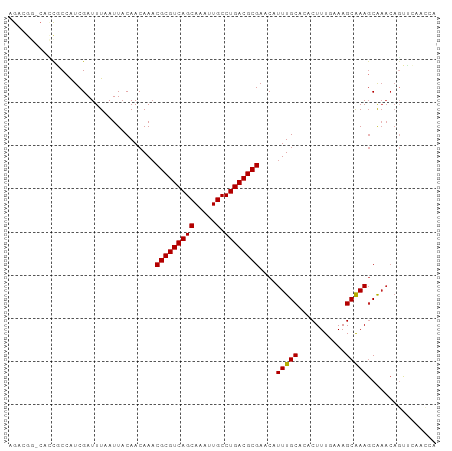
>dm3.chr3L 21783005 98 + 24543557 AGGCGG-CACUGCCAUCGAUUUAAUUACAACAAACGCGUCAGCAAAUUGCCUGACGCGAACAUUUGCACACUUUCGAAGCAAAGCAAACAGUUCAACCA .((..(-.((((......................(((((((((.....).))))))))....(((((...........))))).....)))).)..)). ( -24.70, z-score = -1.98, R) >droSim1.chr3L 21126354 98 + 22553184 AGACGG-CACUGCCAUCGAUUUAAUUACAACAAACGCGUCAGCAAAUUGCCUGACGCGAACAUUUGCACACUUUCGAAGCAAAGCAAACAGUUCAACCA .((.((-(...))).))..................((((((((.....).)))))))((((.(((((................)))))..))))..... ( -22.99, z-score = -2.01, R) >droSec1.super_11 1700720 98 + 2888827 AGACGG-CACUGCCAUCGAUUUAAUUACAACAAACGCGUCAGCAAAUUGCCUGACGCGAACAUUUGCACACUUUCGAAGCAAAGCGAACAGUUCAACCA .(((((-(...))).(((................(((((((((.....).))))))))....(((((...........))))).)))...)))...... ( -23.00, z-score = -1.73, R) >droYak2.chr3L 5478161 98 - 24197627 AGACGG-CACUGCAAUCGAUUUAAUUACAACAAACGCGUCAGCAAAUUGCCUGACGCGAACAUUUGCACACUUUCAAAGCAAAGCAAACAGUUCAACCA ....((-.((((......................(((((((((.....).))))))))....(((((...........))))).....))))....)). ( -21.20, z-score = -1.67, R) >droEre2.scaffold_4784 21455572 98 + 25762168 AGACGG-CACUGCCGUCGAUUUAAUUACAACAAACGCGUCAGCAAAUUGCCUGACGCGAACAUUUGCACACUUUCGAAGCAAAGCAAACAGUUCAGCCA .(((((-(...))))))..................((((((((.....).)))))))((((.(((((................)))))..))))..... ( -28.69, z-score = -2.90, R) >droAna3.scaffold_13337 4095460 84 + 23293914 AGACGG-CAUCGCCAUCGAUUUAAUUACAACAACCGCGUCAGCAAAUUGCCUGACGCGAACAUUUGCACACUUUGAAAGCAAA-CA------------- .((.((-(...))).)).................(((((((((.....).))))))))....(((((...........)))))-..------------- ( -20.90, z-score = -2.43, R) >dp4.chrXR_group6 8890188 99 - 13314419 AGACGGGCAUCGCCAUCGAUUUAAUUACAACAAACGCGUCAGCAAAUUGCCUGACGCGAACAUUUGCACACUUUGAAAGCGAAGCAAACAGUCCAACAA .(((..((.((((..((((...............(((((((((.....).))))))))..............))))..)))).)).....)))...... ( -26.45, z-score = -2.62, R) >droPer1.super_9 918504 99 - 3637205 AGACGGGCAUCGCCAUCGAUUUAAUUACAACAAACGCGUCAGCAAAUUGCCUGACGCGAACAUUUGCACACUUUGAAAGCGAAGCAAACAGUCCAACAA .(((..((.((((..((((...............(((((((((.....).))))))))..............))))..)))).)).....)))...... ( -26.45, z-score = -2.62, R) >droVir3.scaffold_13049 24139301 91 + 25233164 --------AGCUCCAUCGAUUUAAUUACAACAAACGCGUCAGCAAAUUGCCUGACGCGAACAUUUGCACACUUUGAAAGCAAAGCAAACAGCAAAAUCA --------.........(((((............(((((((((.....).))))))))......(((...(((((....)))))......)))))))). ( -21.40, z-score = -2.68, R) >droMoj3.scaffold_6680 18758909 88 + 24764193 --------AGCUCCAUCGAUUUAAUUACAACAAACGCGUCAGCAAAUUGCCUGACGCGAACAUUUGCACACUUUGAAAGCAAAGCAAACACAACCA--- --------.((.......................(((((((((.....).))))))))....(((((...........)))))))...........--- ( -19.30, z-score = -2.76, R) >droGri2.scaffold_15110 14980537 83 - 24565398 --------AGCUCCAUCGAUUUAAUUACAACAAACGCGUCAGCAAAUUGCCUGACGCGAACAUUUGCACACUUUGAAAGCAAAGCAAACAG-------- --------.((.......................(((((((((.....).))))))))....(((((...........)))))))......-------- ( -19.30, z-score = -2.52, R) >consensus AGACGG_CACCGCCAUCGAUUUAAUUACAACAAACGCGUCAGCAAAUUGCCUGACGCGAACAUUUGCACACUUUGAAAGCAAAGCAAACAGUUCAACCA ..................................(((((((((.....).))))))))....(((((...........)))))................ (-17.20 = -17.05 + -0.15)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:42:57 2011