
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 21,703,032 – 21,703,124 |
| Length | 92 |
| Max. P | 0.525807 |

| Location | 21,703,032 – 21,703,124 |
|---|---|
| Length | 92 |
| Sequences | 9 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 61.94 |
| Shannon entropy | 0.74678 |
| G+C content | 0.61240 |
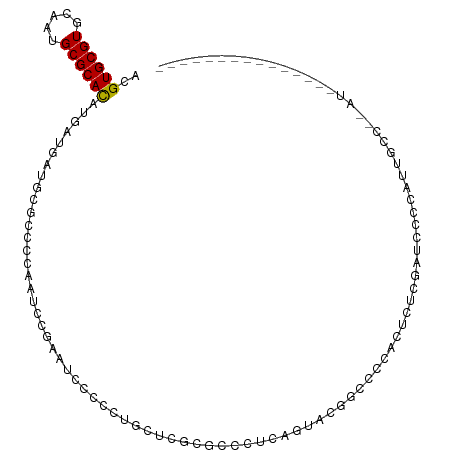
| Mean single sequence MFE | -22.02 |
| Consensus MFE | -9.36 |
| Energy contribution | -9.19 |
| Covariance contribution | -0.17 |
| Combinations/Pair | 1.17 |
| Mean z-score | -0.57 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.525807 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
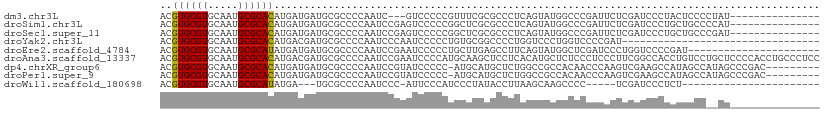
>dm3.chr3L 21703032 92 + 24543557 ACGUGCGUGCAAUGCGCACAUGAUGAUGCGCCCCAAUC---GUCCCCCGUUUCGCGCCCUCAGUAUGGCCCGAUUCUCGAUCCCUACUCCCCUAU--------------- ..((((((.....))))))..((((((........)))---)))....(..(((.(((........))).)))..)...................--------------- ( -22.70, z-score = -1.67, R) >droSim1.chr3L 21042420 95 + 22553184 ACGUGCGUGCAAUGCGCACAUGAUGAUGCGCCCCAAUCCGAGUCCCCCGGCUCGCGCCCUCAGUAUGGCCCGAUUCUCGAUCCCUGCUGCCCCAU--------------- ..((((((.....))))))........((((........(((((....)))))))))...(((((.((..((.....))..)).)))))......--------------- ( -27.10, z-score = -0.64, R) >droSec1.super_11 1623850 95 + 2888827 ACGUGCGUGCAAUGCGCACAUGAUGAUGCGCCCCAAUCCGAGUCCCCCGGCUCGCGCCCUCAGUAUGGCCCGAUUCUCGAUCCCUGCUGCCCGAU--------------- .((.((((((...((((((.....).)))))........(((((....)))))))))....((((.((..((.....))..)).)))))).))..--------------- ( -28.50, z-score = -0.74, R) >droYak2.chr3L 5389514 77 - 24197627 ACGUGCGUGCAAUGCGCACAUGACGAUGCGCCCCAAUCCCAAUCCCCCUGUGCGGCCCCUGGUCCCUGGUCCCCGAU--------------------------------- ..((((((.....))))))....((..((((..................))))((..((.(....).))..))))..--------------------------------- ( -17.77, z-score = 0.93, R) >droEre2.scaffold_4784 21360041 88 + 25762168 ACGUGCGUGCAAUGCGCAUAUGAUGAUGCGCCCCAAUCCGAAUCCCCCUGCUUGAGCCUUCAGUAUGGCUCGAUCCCUGGUCCCCGAU---------------------- .((.(.(..(...((((((......))))))....................(((((((........)))))))......)..))))..---------------------- ( -20.90, z-score = -0.13, R) >droAna3.scaffold_13337 4024868 110 + 23293914 ACGUGCGUGCAAUGCGCACAUGACGAUGCGCCCCAAUCCGAAUCCCCAUGCAAGCUCCUCACAUGCUCUCCCUCCCUUCGGCCACCUGUCCUGCUCCCCACCUGCCCUCC ..((((((.....))))))..((((.((.(((..............((((..((...))..))))..............)))))..)))).................... ( -18.29, z-score = -0.16, R) >dp4.chrXR_group6 8813760 100 - 13314419 ACGUGCGUGCAAUGCGCACAUGAUGAUGCGCCCCAAUCCGUAUCCCCC-AUGCAUGCUCUGGCCGCCACAACCCAAGUCGAAGCCAUAGCCAUAGCCCGAC--------- ..((((((.....))))))(((..((((((........))))))...)-))(((((((.((((..(.((.......)).)..)))).)).))).)).....--------- ( -26.10, z-score = -0.94, R) >droPer1.super_9 842442 100 - 3637205 ACGUGCGUGCAAUGCGCACAUGAUGAUGCGCCCCAAUCCGUAUCCCCC-AUGCAUGCUCUGGCCGCCACAACCCAAGUCGAAGCCAUAGCCAUAGCCCGAC--------- ..((((((.....))))))(((..((((((........))))))...)-))(((((((.((((..(.((.......)).)..)))).)).))).)).....--------- ( -26.10, z-score = -0.94, R) >droWil1.scaffold_180698 10378528 78 - 11422946 ACGUGCGUGCAAUGCGCAUAUGA---UGCGCCCCAAUCCC-AUUCCCAUCCCUAUACCUUAAGCAAGCCCC-----UCGAUCCCUCU----------------------- .((.((.(((...((((((...)---))))).........-...........((.....)).))).))...-----.))........----------------------- ( -10.70, z-score = -0.81, R) >consensus ACGUGCGUGCAAUGCGCACAUGAUGAUGCGCCCCAAUCCGAAUCCCCCUGCUCGCGCCCUCAGUACGGCCCCACUCUCGAUCCCCAUUGCC__AU_______________ ..((((((.....))))))........................................................................................... ( -9.36 = -9.19 + -0.17)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:42:53 2011