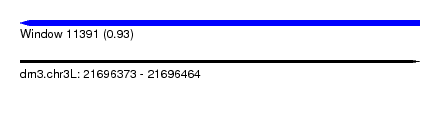
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 21,696,373 – 21,696,464 |
| Length | 91 |
| Max. P | 0.925842 |

| Location | 21,696,373 – 21,696,464 |
|---|---|
| Length | 91 |
| Sequences | 8 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 66.82 |
| Shannon entropy | 0.63419 |
| G+C content | 0.28523 |
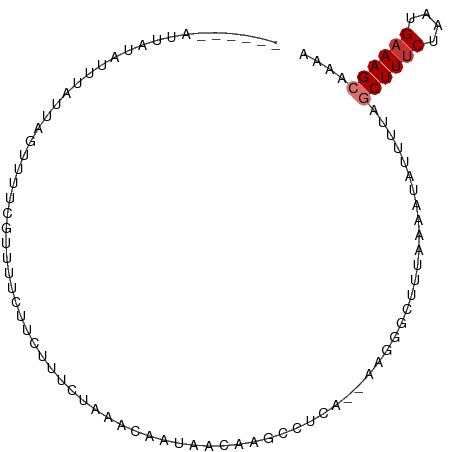
| Mean single sequence MFE | -15.87 |
| Consensus MFE | -5.72 |
| Energy contribution | -6.10 |
| Covariance contribution | 0.38 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.36 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.32 |
| SVM RNA-class probability | 0.925842 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr3L 21696373 91 - 24543557 ------AUUAUUUUUAUUAGUUUUCCAUUUCUUCUUUCUACAUAACAAGAAGACUCA--AAGGGCUUUAAACUAUUUUAGCUUUCUAAUGAAAGCACAA ------...........((((((.((...(((((((..........)))))))....--...))....)))))).....((((((....)))))).... ( -16.00, z-score = -2.81, R) >droSim1.chr3L 21035677 91 - 22553184 ------AUUAUAUUUAUUAGUUUUCGUUUUCUUCUUUCUAAACAAUAACAAGGCUCA--AAGGGCUCUAAACUAUUUUAGCUUUCUAAUGAAAGCACAA ------...........((((((..((((..........)))).......(((((..--...))).)))))))).....((((((....)))))).... ( -14.90, z-score = -2.37, R) >droSec1.super_11 1617213 91 - 2888827 ------AUUAUAUUUAUUAGUUUUCGUUUUCUUCUUUCUAAACAAUAACAAGGCUCA--AAGGGCUCUAAACUAUUUUAGCUUUCUAAUGAAAGCACAA ------...........((((((..((((..........)))).......(((((..--...))).)))))))).....((((((....)))))).... ( -14.90, z-score = -2.37, R) >droEre2.scaffold_4784 21353340 89 - 25762168 ------AAUUUAUUUAUUAGUUUUCGUUUUCUUCUUGCUAAACAACAAGAAGACUUA--A--GCCUUAAAAAUAUUUUGGCUUUCUAAUGAAAGCACAA ------.............(((((((((.((((((((........))))))))...(--(--(((..((.....))..)))))...))))))))).... ( -21.50, z-score = -3.89, R) >dp4.chrXR_group6 8807024 96 + 13314419 AGUAUGAUUAUGAUUAUUAGUUUUUGUCUCUUUUCUAAACAAUAAUAAAAAGCCCUA--AAAGGCUUUCAAA-ACUUCAGCUUUCUAAUGAAAGGAAAA ....(((.....((((((.((((.............))))))))))..((((((...--...))))))....-...))).(((((....)))))..... ( -13.32, z-score = -0.05, R) >droPer1.super_9 835645 96 + 3637205 AGUAUGAUUAUGAUUAUUAGUUUUUGUCUCUUUUCUAAACAAUAAUAAAAAGCCCUA--AAAGGCUUUCAAA-ACUUCAGCUUUCUAAUGAAAGGAAAA ....(((.....((((((.((((.............))))))))))..((((((...--...))))))....-...))).(((((....)))))..... ( -13.32, z-score = -0.05, R) >droMoj3.scaffold_6680 5909222 76 + 24764193 --------------CAAUG--UUUUUGUUUAGCUUUUCUAAACA--AACACACAACA--AAGGCCUUU---GUACGCUCGCUUUCUAAUGCAAGCAACA --------------...((--(.(((((((((.....)))))))--)).)))..(((--((....)))---))..(((.((........)).))).... ( -17.50, z-score = -2.74, R) >droGri2.scaffold_15110 9515007 81 - 24565398 --------------CAAUAUUUUUUUGUUUAACUUUUCUAAACAACAACAGCCAGCACCAAAGCCUUU----UAUGCUGGCUUUCUAAUGAAAGCAAAA --------------..........(((((((.......))))))).......(((((...........----..)))))((((((....)))))).... ( -15.52, z-score = -1.69, R) >consensus ______AUUAUAUUUAUUAGUUUUCGUUUUCUUCUUUCUAAACAAUAACAAGCCUCA__AAGGGCUUUAAAAUAUUUUAGCUUUCUAAUGAAAGCAAAA ...............................................................................((((((....)))))).... ( -5.72 = -6.10 + 0.38)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:42:52 2011