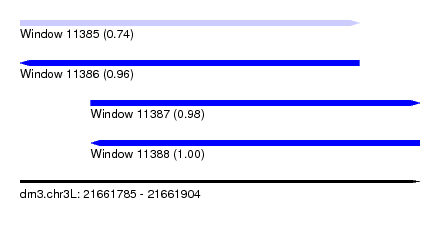
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 21,661,785 – 21,661,904 |
| Length | 119 |
| Max. P | 0.998955 |

| Location | 21,661,785 – 21,661,886 |
|---|---|
| Length | 101 |
| Sequences | 11 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 79.87 |
| Shannon entropy | 0.40867 |
| G+C content | 0.41757 |
| Mean single sequence MFE | -21.25 |
| Consensus MFE | -16.92 |
| Energy contribution | -16.92 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -0.97 |
| Structure conservation index | 0.80 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.742024 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 21661785 101 + 24543557 ----------CACCUGGAUCACCUGCAAAUCGCUGAGCAAACUCAUAGUAAAUACGCCUUUUUCCCUUUCCUGUCGUUGUGGAAUUGAAACGCCAGUAAGCGGAAAAAGAC ----------..............((.......((((....))))..((....))))(((((((((((..(((.((((..........)))).))).))).)))))))).. ( -19.90, z-score = -0.08, R) >droPer1.super_9 791477 98 - 3637205 -------------GAAUUGGGUCGGUUGUUUGUUGGAUAAACUCAUAGUAAAUACGCCUUUUUCCCUUUCCUGUCGUUGAUCGAAUAAAACGCCAGUAAGCGGAAAAAGAC -------------....(((((..(((........)))..)))))............(((((((((((..(((.((((..........)))).))).))).)))))))).. ( -21.30, z-score = -0.42, R) >dp4.chrXR_group6 8765729 98 - 13314419 -------------GAAUUGGGUCGGUUGUUUGUUGGAUAAACUCAUAGUAAAUACGCCUUUUUCCCUUUCCUGUCGUUGAUCGAAUAAAACGCCAGUAAGCGGAAAAAGAC -------------....(((((..(((........)))..)))))............(((((((((((..(((.((((..........)))).))).))).)))))))).. ( -21.30, z-score = -0.42, R) >droAna3.scaffold_13337 3985733 94 + 23293914 --------------GACUUUGACGAAA---UGUUGACCAAACUCAUAGUAAAUACGCCUUUUUCCCUUUCCUGUCGUUGUUUGAAUUAAACGCCAGUAAGCGGAAAAAGAC --------------.(((.(((.....---.(((.....)))))).)))........(((((((((((..(((.((((..........)))).))).))).)))))))).. ( -18.90, z-score = -1.57, R) >droEre2.scaffold_4784 21318794 101 + 25762168 ----------CACCUUGAUCACCUGCAAGUCGCUGGCCAAACUCAUAGUGAAUACGCCUUUUUCCCUUUCCUGUCGUUGUUGAAAUGAAACGCCAGUAAGCGGAAAAAGAC ----------(((..(((...((.((.....)).))......)))..))).......(((((((((((..(((.((((..........)))).))).))).)))))))).. ( -23.70, z-score = -1.46, R) >droYak2.chr3L 5348001 101 - 24197627 ----------CACCUUGAUCACCUGCAAAUCGCUGGUCAAACUCAUAGUAAAUACGCCUUUUUCCCUUUCCUGUCGUUGUUGAAAUGAAACGCCAGUAAGCGGAAAAAGAC ----------....(((((((...((.....))))))))).................(((((((((((..(((.((((..........)))).))).))).)))))))).. ( -23.40, z-score = -2.15, R) >droSec1.super_11 1581853 101 + 2888827 ----------CACCUGGAUCACCUGCAAGUCGCUGGGCAAACUCAUAGUAAAUACGCCUUUUUCCCUUUCCUGUCGUUGUGGAAAUGAAACGCCAGUAAGCGGAAAAAGAC ----------..((..(...((......))..)..))....................(((((((((((..(((.((((..........)))).))).))).)))))))).. ( -21.30, z-score = 0.36, R) >droSim1.chr3L 21004812 101 + 22553184 ----------CACCUGGAUCACCUGCAAGUCGCUGGGCAAACUCAUAGUAAAUACGCCUUUUUCCCUUUCCUGUCGUUGUGGAAAUGAAACGCCAGUAAGCGGAAAAAGAC ----------..((..(...((......))..)..))....................(((((((((((..(((.((((..........)))).))).))).)))))))).. ( -21.30, z-score = 0.36, R) >droWil1.scaffold_180698 10330114 111 - 11422946 UACUUAGUUGUAGAGAUUUUGUGGAGAAUUUGUUGAUUAAACUCAUAGUAAAUACGCCUUUUUCCCUUUCCUGUCGUUAGUUAAAUAAAACGCCAGUAAGCGGAAAAAGAC ((((.((((.((((((((((....))))))).)))....))))...)))).......(((((((((((..(((.((((..........)))).))).))).)))))))).. ( -21.30, z-score = -0.92, R) >droVir3.scaffold_13049 966216 94 - 25233164 --------------UGUUUAAACAG--AUUGGUUGAACAAAUUCAUAGUAAAUACGCCUUUUUCCCUUUCCUGUCGUUGCUC-AAUAAAACGCCAGUAAGCGGAAAAAGAC --------------(((((((.(..--...).)))))))..................(((((((((((..(((.((((....-.....)))).))).))).)))))))).. ( -20.90, z-score = -2.22, R) >droGri2.scaffold_15110 9482030 94 + 24565398 --------------UAUUUGAAUAG--GUUUGUUGAUUACAUUCAUAGUAAAUACGCCUUUUUCCCUUUCCUGUCGUUGCUC-AAUAAAACGCCAGUAAGCGGAAAAAGAC --------------((((((.....--(..(((.....)))..)....))))))...(((((((((((..(((.((((....-.....)))).))).))).)))))))).. ( -20.40, z-score = -2.08, R) >consensus ___________A_CUAGUUCACCUGCAAUUCGUUGGACAAACUCAUAGUAAAUACGCCUUUUUCCCUUUCCUGUCGUUGUUGAAAUAAAACGCCAGUAAGCGGAAAAAGAC .........................................................(((((((((((..(((.((((..........)))).))).))).)))))))).. (-16.92 = -16.92 + -0.00)

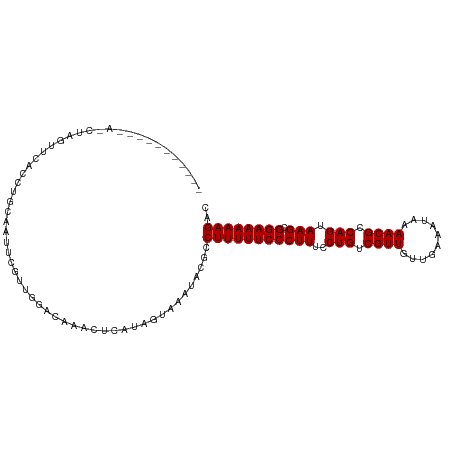
| Location | 21,661,785 – 21,661,886 |
|---|---|
| Length | 101 |
| Sequences | 11 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 79.87 |
| Shannon entropy | 0.40867 |
| G+C content | 0.41757 |
| Mean single sequence MFE | -24.83 |
| Consensus MFE | -20.76 |
| Energy contribution | -20.76 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.84 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.70 |
| SVM RNA-class probability | 0.961672 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 21661785 101 - 24543557 GUCUUUUUCCGCUUACUGGCGUUUCAAUUCCACAACGACAGGAAAGGGAAAAAGGCGUAUUUACUAUGAGUUUGCUCAGCGAUUUGCAGGUGAUCCAGGUG---------- ((((((((((.(((.(((.((((..........)))).)))..)))))))))))))....(((((.((((....))))((.....)).)))))........---------- ( -28.10, z-score = -1.25, R) >droPer1.super_9 791477 98 + 3637205 GUCUUUUUCCGCUUACUGGCGUUUUAUUCGAUCAACGACAGGAAAGGGAAAAAGGCGUAUUUACUAUGAGUUUAUCCAACAAACAACCGACCCAAUUC------------- ((((((((((.(((.(((.((((..........)))).)))..))))))))))))).............((((.......))))..............------------- ( -21.00, z-score = -1.21, R) >dp4.chrXR_group6 8765729 98 + 13314419 GUCUUUUUCCGCUUACUGGCGUUUUAUUCGAUCAACGACAGGAAAGGGAAAAAGGCGUAUUUACUAUGAGUUUAUCCAACAAACAACCGACCCAAUUC------------- ((((((((((.(((.(((.((((..........)))).)))..))))))))))))).............((((.......))))..............------------- ( -21.00, z-score = -1.21, R) >droAna3.scaffold_13337 3985733 94 - 23293914 GUCUUUUUCCGCUUACUGGCGUUUAAUUCAAACAACGACAGGAAAGGGAAAAAGGCGUAUUUACUAUGAGUUUGGUCAACA---UUUCGUCAAAGUC-------------- ((((((((((.(((.(((.((((..........)))).)))..)))))))))))))...........((.(((((.(....---....)))))).))-------------- ( -23.50, z-score = -2.31, R) >droEre2.scaffold_4784 21318794 101 - 25762168 GUCUUUUUCCGCUUACUGGCGUUUCAUUUCAACAACGACAGGAAAGGGAAAAAGGCGUAUUCACUAUGAGUUUGGCCAGCGACUUGCAGGUGAUCAAGGUG---------- ((((((((((.(((.(((.((((..........)))).)))..))))))))))))).....((((.(((...(.(((.((.....)).))).)))).))))---------- ( -29.80, z-score = -1.71, R) >droYak2.chr3L 5348001 101 + 24197627 GUCUUUUUCCGCUUACUGGCGUUUCAUUUCAACAACGACAGGAAAGGGAAAAAGGCGUAUUUACUAUGAGUUUGACCAGCGAUUUGCAGGUGAUCAAGGUG---------- ((((((((((.(((.(((.((((..........)))).)))..))))))))))))).....((((.(((...(.(((.((.....)).))).)))).))))---------- ( -28.00, z-score = -1.71, R) >droSec1.super_11 1581853 101 - 2888827 GUCUUUUUCCGCUUACUGGCGUUUCAUUUCCACAACGACAGGAAAGGGAAAAAGGCGUAUUUACUAUGAGUUUGCCCAGCGACUUGCAGGUGAUCCAGGUG---------- ((((((((((.(((.(((.((((..........)))).)))..))))))))))))).....((((.((.(.(..((..((.....)).))..).)))))))---------- ( -27.10, z-score = -0.66, R) >droSim1.chr3L 21004812 101 - 22553184 GUCUUUUUCCGCUUACUGGCGUUUCAUUUCCACAACGACAGGAAAGGGAAAAAGGCGUAUUUACUAUGAGUUUGCCCAGCGACUUGCAGGUGAUCCAGGUG---------- ((((((((((.(((.(((.((((..........)))).)))..))))))))))))).....((((.((.(.(..((..((.....)).))..).)))))))---------- ( -27.10, z-score = -0.66, R) >droWil1.scaffold_180698 10330114 111 + 11422946 GUCUUUUUCCGCUUACUGGCGUUUUAUUUAACUAACGACAGGAAAGGGAAAAAGGCGUAUUUACUAUGAGUUUAAUCAACAAAUUCUCCACAAAAUCUCUACAACUAAGUA ((((((((((.(((.(((.((((..........)))).)))..)))))))))))))...........((((((.......))))))......................... ( -23.20, z-score = -2.50, R) >droVir3.scaffold_13049 966216 94 + 25233164 GUCUUUUUCCGCUUACUGGCGUUUUAUU-GAGCAACGACAGGAAAGGGAAAAAGGCGUAUUUACUAUGAAUUUGUUCAACCAAU--CUGUUUAAACA-------------- ((((((((((.(((.(((.((((.....-....)))).)))..)))))))))))))..........((((....))))......--...........-------------- ( -21.80, z-score = -1.50, R) >droGri2.scaffold_15110 9482030 94 - 24565398 GUCUUUUUCCGCUUACUGGCGUUUUAUU-GAGCAACGACAGGAAAGGGAAAAAGGCGUAUUUACUAUGAAUGUAAUCAACAAAC--CUAUUCAAAUA-------------- ((((((((((.(((.(((.((((.....-....)))).)))..)))))))))))))((..((((.......))))...))....--...........-------------- ( -22.50, z-score = -2.37, R) >consensus GUCUUUUUCCGCUUACUGGCGUUUCAUUUCAACAACGACAGGAAAGGGAAAAAGGCGUAUUUACUAUGAGUUUGACCAACAAAUUGCAGGUGAAAUAG_U___________ ((((((((((.(((.(((.((((..........)))).)))..)))))))))))))....................................................... (-20.76 = -20.76 + 0.00)

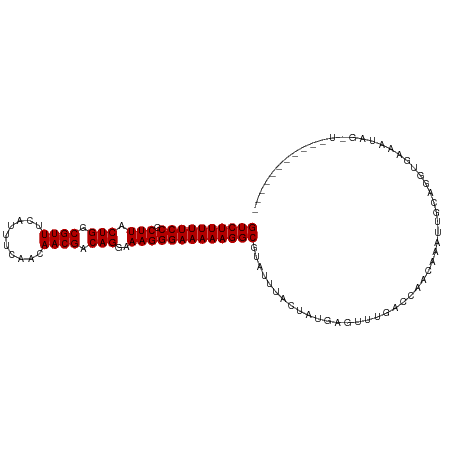
| Location | 21,661,806 – 21,661,904 |
|---|---|
| Length | 98 |
| Sequences | 12 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 85.53 |
| Shannon entropy | 0.29022 |
| G+C content | 0.38511 |
| Mean single sequence MFE | -22.42 |
| Consensus MFE | -19.16 |
| Energy contribution | -19.16 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.85 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.97 |
| SVM RNA-class probability | 0.977079 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 21661806 98 + 24543557 GCUGAGCAAACUCAUAGUAAAUACGCCUUUUUCCCUUUCCUGUCGUUGUGGAAUUGAAACGCCAGUAAGCGGAAAAAGACAAAUCGACUAGAAUUUGU------------------ .....(((((.((.((((........(((((((((((..(((.((((..........)))).))).))).))))))))........)))))).)))))------------------ ( -23.79, z-score = -1.98, R) >droPer1.super_9 791495 102 - 3637205 GUUGGAUAAACUCAUAGUAAAUACGCCUUUUUCCCUUUCCUGUCGUUGAUCGAAUAAAACGCCAGUAAGCGGAAAAAGACAAAUCGACUAGAAUUUUGAUCU-------------- ...((((.......((((........(((((((((((..(((.((((..........)))).))).))).))))))))........))))........))))-------------- ( -21.25, z-score = -1.63, R) >dp4.chrXR_group6 8765747 102 - 13314419 GUUGGAUAAACUCAUAGUAAAUACGCCUUUUUCCCUUUCCUGUCGUUGAUCGAAUAAAACGCCAGUAAGCGGAAAAAGACAAAUCGACUAGAAUUUUGAUCU-------------- ...((((.......((((........(((((((((((..(((.((((..........)))).))).))).))))))))........))))........))))-------------- ( -21.25, z-score = -1.63, R) >droAna3.scaffold_13337 3985747 98 + 23293914 GUUGACCAAACUCAUAGUAAAUACGCCUUUUUCCCUUUCCUGUCGUUGUUUGAAUUAAACGCCAGUAAGCGGAAAAAGACAAAUCGACUAGAAUUUGA------------------ ......((((.((.((((........(((((((((((..(((.((((..........)))).))).))).))))))))........)))))).)))).------------------ ( -22.89, z-score = -2.90, R) >droEre2.scaffold_4784 21318815 98 + 25762168 GCUGGCCAAACUCAUAGUGAAUACGCCUUUUUCCCUUUCCUGUCGUUGUUGAAAUGAAACGCCAGUAAGCGGAAAAAGACAAAUCGACUAGAAUUUGU------------------ ......((((.((.((((((....(.(((((((((((..(((.((((..........)))).))).))).)))))))).)...)).)))))).)))).------------------ ( -21.90, z-score = -1.39, R) >droYak2.chr3L 5348022 98 - 24197627 GCUGGUCAAACUCAUAGUAAAUACGCCUUUUUCCCUUUCCUGUCGUUGUUGAAAUGAAACGCCAGUAAGCGGAAAAAGACAAAUCGACUAGAAUUUGU------------------ .((((((.........((....))(.(((((((((((..(((.((((..........)))).))).))).)))))))).).....)))))).......------------------ ( -24.70, z-score = -2.70, R) >droSec1.super_11 1581874 98 + 2888827 GCUGGGCAAACUCAUAGUAAAUACGCCUUUUUCCCUUUCCUGUCGUUGUGGAAAUGAAACGCCAGUAAGCGGAAAAAGACAAAUCGACUAGAAUUUGU------------------ .....(((((.((.((((........(((((((((((..(((.((((..........)))).))).))).))))))))........)))))).)))))------------------ ( -23.89, z-score = -1.49, R) >droSim1.chr3L 21004833 98 + 22553184 GCUGGGCAAACUCAUAGUAAAUACGCCUUUUUCCCUUUCCUGUCGUUGUGGAAAUGAAACGCCAGUAAGCGGAAAAAGACAAAUCGACUAGAAUUUGU------------------ .....(((((.((.((((........(((((((((((..(((.((((..........)))).))).))).))))))))........)))))).)))))------------------ ( -23.89, z-score = -1.49, R) >droWil1.scaffold_180698 10330145 98 - 11422946 GUUGAUUAAACUCAUAGUAAAUACGCCUUUUUCCCUUUCCUGUCGUUAGUUAAAUAAAACGCCAGUAAGCGGAAAAAGACAAAUCGACUAGAAUUUAU------------------ (((((((.........((....))(.(((((((((((..(((.((((..........)))).))).))).)))))))).).)))))))..........------------------ ( -21.70, z-score = -3.39, R) >droVir3.scaffold_13049 966231 97 - 25233164 GUUGAACAAAUUCAUAGUAAAUACGCCUUUUUCCCUUUCCUGUCGUU-GCUCAAUAAAACGCCAGUAAGCGGAAAAAGACAAAUCGACUAGAACUUUA------------------ (((((...........((....))(.(((((((((((..(((.((((-.........)))).))).))).)))))))).)...)))))..........------------------ ( -20.80, z-score = -2.80, R) >droMoj3.scaffold_6680 5872890 115 - 24764193 GUUGAAAUAAUUCAUAGUAAAUACGCCUUUUUCCCUUCCCUGCCGUU-GUUCAAUAAAACGUCAGUAAGCGGAAAAAGACAAAUCGACUAGAACUUUAUAUAAAAAUCGAUUCAGU (((((...........((....))(.(((((((((((..(((.((((-.........)))).))).))).)))))))).)...)))))............................ ( -20.30, z-score = -2.07, R) >droGri2.scaffold_15110 9482045 105 + 24565398 GUUGAUUACAUUCAUAGUAAAUACGCCUUUUUCCCUUUCCUGUCGUU-GCUCAAUAAAACGCCAGUAAGCGGAAAAAGACAAAUCGACUAGAACUCUG----AAGAUUGC------ (((((((.........((....))(.(((((((((((..(((.((((-.........)))).))).))).)))))))).).)))))))..........----........------ ( -22.70, z-score = -2.07, R) >consensus GUUGAACAAACUCAUAGUAAAUACGCCUUUUUCCCUUUCCUGUCGUUGUUGAAAUAAAACGCCAGUAAGCGGAAAAAGACAAAUCGACUAGAAUUUGU__________________ ...........((.((((........(((((((((((..(((.((((..........)))).))).))).))))))))........))))))........................ (-19.16 = -19.16 + 0.00)
| Location | 21,661,806 – 21,661,904 |
|---|---|
| Length | 98 |
| Sequences | 12 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 85.53 |
| Shannon entropy | 0.29022 |
| G+C content | 0.38511 |
| Mean single sequence MFE | -31.55 |
| Consensus MFE | -31.16 |
| Energy contribution | -31.03 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.14 |
| Mean z-score | -4.11 |
| Structure conservation index | 0.99 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.57 |
| SVM RNA-class probability | 0.998955 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 21661806 98 - 24543557 ------------------ACAAAUUCUAGUCGAUUUGUCUUUUUCCGCUUACUGGCGUUUCAAUUCCACAACGACAGGAAAGGGAAAAAGGCGUAUUUACUAUGAGUUUGCUCAGC ------------------.(((((((((((.(((.(((((((((((.(((.(((.((((..........)))).)))..)))))))))))))).))).)))).)))))))...... ( -32.50, z-score = -4.36, R) >droPer1.super_9 791495 102 + 3637205 --------------AGAUCAAAAUUCUAGUCGAUUUGUCUUUUUCCGCUUACUGGCGUUUUAUUCGAUCAACGACAGGAAAGGGAAAAAGGCGUAUUUACUAUGAGUUUAUCCAAC --------------.(((..((((((((((.(((.(((((((((((.(((.(((.((((..........)))).)))..)))))))))))))).))).)))).))))))))).... ( -30.20, z-score = -3.63, R) >dp4.chrXR_group6 8765747 102 + 13314419 --------------AGAUCAAAAUUCUAGUCGAUUUGUCUUUUUCCGCUUACUGGCGUUUUAUUCGAUCAACGACAGGAAAGGGAAAAAGGCGUAUUUACUAUGAGUUUAUCCAAC --------------.(((..((((((((((.(((.(((((((((((.(((.(((.((((..........)))).)))..)))))))))))))).))).)))).))))))))).... ( -30.20, z-score = -3.63, R) >droAna3.scaffold_13337 3985747 98 - 23293914 ------------------UCAAAUUCUAGUCGAUUUGUCUUUUUCCGCUUACUGGCGUUUAAUUCAAACAACGACAGGAAAGGGAAAAAGGCGUAUUUACUAUGAGUUUGGUCAAC ------------------((((((((((((.(((.(((((((((((.(((.(((.((((..........)))).)))..)))))))))))))).))).)))).))))))))..... ( -33.20, z-score = -5.23, R) >droEre2.scaffold_4784 21318815 98 - 25762168 ------------------ACAAAUUCUAGUCGAUUUGUCUUUUUCCGCUUACUGGCGUUUCAUUUCAACAACGACAGGAAAGGGAAAAAGGCGUAUUCACUAUGAGUUUGGCCAGC ------------------.(((((((((((.(((.(((((((((((.(((.(((.((((..........)))).)))..)))))))))))))).))).)))).)))))))...... ( -33.40, z-score = -4.46, R) >droYak2.chr3L 5348022 98 + 24197627 ------------------ACAAAUUCUAGUCGAUUUGUCUUUUUCCGCUUACUGGCGUUUCAUUUCAACAACGACAGGAAAGGGAAAAAGGCGUAUUUACUAUGAGUUUGACCAGC ------------------.(((((((((((.(((.(((((((((((.(((.(((.((((..........)))).)))..)))))))))))))).))).)))).)))))))...... ( -33.20, z-score = -4.79, R) >droSec1.super_11 1581874 98 - 2888827 ------------------ACAAAUUCUAGUCGAUUUGUCUUUUUCCGCUUACUGGCGUUUCAUUUCCACAACGACAGGAAAGGGAAAAAGGCGUAUUUACUAUGAGUUUGCCCAGC ------------------.(((((((((((.(((.(((((((((((.(((.(((.((((..........)))).)))..)))))))))))))).))).)))).)))))))...... ( -32.50, z-score = -4.19, R) >droSim1.chr3L 21004833 98 - 22553184 ------------------ACAAAUUCUAGUCGAUUUGUCUUUUUCCGCUUACUGGCGUUUCAUUUCCACAACGACAGGAAAGGGAAAAAGGCGUAUUUACUAUGAGUUUGCCCAGC ------------------.(((((((((((.(((.(((((((((((.(((.(((.((((..........)))).)))..)))))))))))))).))).)))).)))))))...... ( -32.50, z-score = -4.19, R) >droWil1.scaffold_180698 10330145 98 + 11422946 ------------------AUAAAUUCUAGUCGAUUUGUCUUUUUCCGCUUACUGGCGUUUUAUUUAACUAACGACAGGAAAGGGAAAAAGGCGUAUUUACUAUGAGUUUAAUCAAC ------------------.(((((((((((.(((.(((((((((((.(((.(((.((((..........)))).)))..)))))))))))))).))).)))).)))))))...... ( -32.10, z-score = -5.44, R) >droVir3.scaffold_13049 966231 97 + 25233164 ------------------UAAAGUUCUAGUCGAUUUGUCUUUUUCCGCUUACUGGCGUUUUAUUGAGC-AACGACAGGAAAGGGAAAAAGGCGUAUUUACUAUGAAUUUGUUCAAC ------------------..((((((((((.(((.(((((((((((.(((.(((.((((.........-)))).)))..)))))))))))))).))).)))).))))))....... ( -30.10, z-score = -3.86, R) >droMoj3.scaffold_6680 5872890 115 + 24764193 ACUGAAUCGAUUUUUAUAUAAAGUUCUAGUCGAUUUGUCUUUUUCCGCUUACUGACGUUUUAUUGAAC-AACGGCAGGGAAGGGAAAAAGGCGUAUUUACUAUGAAUUAUUUCAAC ...(((((((((...((.....))...)))))))))((((((((((.(((.(((.((((.........-)))).)))..)))))))))))))..........((((....)))).. ( -30.10, z-score = -3.18, R) >droGri2.scaffold_15110 9482045 105 - 24565398 ------GCAAUCUU----CAGAGUUCUAGUCGAUUUGUCUUUUUCCGCUUACUGGCGUUUUAUUGAGC-AACGACAGGAAAGGGAAAAAGGCGUAUUUACUAUGAAUGUAAUCAAC ------........----....((((((((.(((.(((((((((((.(((.(((.((((.........-)))).)))..)))))))))))))).))).)))).))))......... ( -28.60, z-score = -2.40, R) >consensus __________________ACAAAUUCUAGUCGAUUUGUCUUUUUCCGCUUACUGGCGUUUUAUUUAAACAACGACAGGAAAGGGAAAAAGGCGUAUUUACUAUGAGUUUGUCCAAC ...................(((((((((((.(((.(((((((((((.(((.(((.((((..........)))).)))..)))))))))))))).))).)))).)))))))...... (-31.16 = -31.03 + -0.12)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:42:49 2011