
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 21,656,196 – 21,656,283 |
| Length | 87 |
| Max. P | 0.967656 |

| Location | 21,656,196 – 21,656,283 |
|---|---|
| Length | 87 |
| Sequences | 7 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 73.07 |
| Shannon entropy | 0.53662 |
| G+C content | 0.67294 |
| Mean single sequence MFE | -39.40 |
| Consensus MFE | -17.58 |
| Energy contribution | -19.07 |
| Covariance contribution | 1.49 |
| Combinations/Pair | 1.30 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.79 |
| SVM RNA-class probability | 0.967656 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
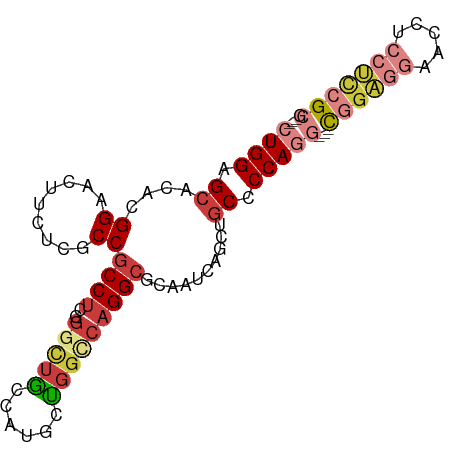
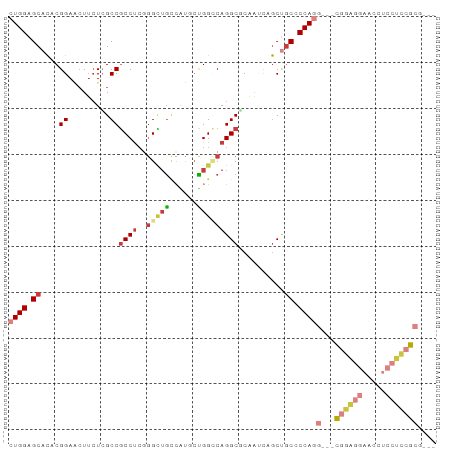
>dm3.chr3L 21656196 87 + 24543557 CUGGAGCACACGGAACUUCUCGCCGCCUCCGGCUGCCAUGCUGGCCAGGCGCAAUCAGCUGCUCCAGG---CGGAGGAACCUCCUCCGCG--- (((((((((...((.....))((.(((((((((......)))))..)))))).....).))))))))(---((((((.....))))))).--- ( -44.90, z-score = -3.63, R) >droAna3.scaffold_13337 3980390 93 + 23293914 CUGGAGCAUACGGAGCUGCUAGCCGCCUCCGGUUGUGGCUCCGGACAGGCUCCUCCUACAGCACCACAGACUGCCGGAGCAGGAGCAGCACCC ..((.((...((((((..(.(((((....))))))..)))))).....((((((.((.(.(((........))).).)).)))))).)).)). ( -41.00, z-score = -1.54, R) >droWil1.scaffold_180698 10321159 84 - 11422946 CUGGAGCACACGGAAAUUAUGGCCACCCAGAGUACAACUCCGGGUCAGGCUCCU---GCCUC-CCAUGGGCCAGCCAAUUCCCAUUCG----- ...........((((....(((((((((.(((.....))).)))).((((....---)))).-.....))))).....))))......----- ( -27.90, z-score = -1.34, R) >droSec1.super_11 1576113 87 + 2888827 CUGGAGCACACGGAACUUCUCGCCGCCUCGGGAUGCCAUGCUGGCCAGGCGCAAUCAGCUGCCCCAGG---CGGAGGAACUUCCUCCGCG--- ((((((((...((....(((((......)))))..)).))))(((..(((.......))))))))))(---((((((.....))))))).--- ( -36.40, z-score = -1.01, R) >droSim1.chr3L 20999155 87 + 22553184 CUGGAGCACACGGAACUUCUCGCCGCCUCGGGCUGCCAUGGUGGCCAGGCGCAAUCAGCUGCCCCAGG---CGGAGGAACUUCCUCCGCG--- ((((.((((...((.....))((.((((..(((..(....)..))))))))).....).))).))))(---((((((.....))))))).--- ( -41.70, z-score = -2.03, R) >droEre2.scaffold_4784 21312719 87 + 25762168 CUGGAGCACACGGAACUUCUCGCCGCCUCGGGCUGCCAUUCCGGCCAGGCCCAAUCCGCUGCCCCAGG---CGGAGGAACCUCCUCCGCG--- ((((.(((..((((.......(......)((((((((.....)))..)))))..)))).))).))))(---((((((.....))))))).--- ( -40.70, z-score = -2.53, R) >droYak2.chr3L 5341880 84 - 24197627 CUGGAGCACACGGAACUUCUCGCCGCCUCGGGCUGCCAUUCUGGGCAGGCUCAA---GCUGCUCCAGG---CGGAGGAACCUCCUCCGCG--- (((((((((.(((.........)))....((((((((......)))).))))..---).))))))))(---((((((.....))))))).--- ( -43.20, z-score = -3.37, R) >consensus CUGGAGCACACGGAACUUCUCGCCGCCUCGGGCUGCCAUGCUGGCCAGGCGCAAUCAGCUGCCCCAGG___CGGAGGAACCUCCUCCGCG___ ((((.((....((.........))((((..(((((......)))))))))..........)).))))....((((((.....))))))..... (-17.58 = -19.07 + 1.49)

| Location | 21,656,196 – 21,656,283 |
|---|---|
| Length | 87 |
| Sequences | 7 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 73.07 |
| Shannon entropy | 0.53662 |
| G+C content | 0.67294 |
| Mean single sequence MFE | -42.97 |
| Consensus MFE | -19.10 |
| Energy contribution | -20.17 |
| Covariance contribution | 1.07 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.24 |
| SVM RNA-class probability | 0.914406 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
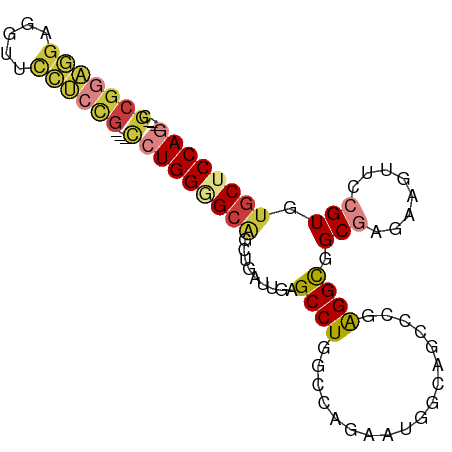
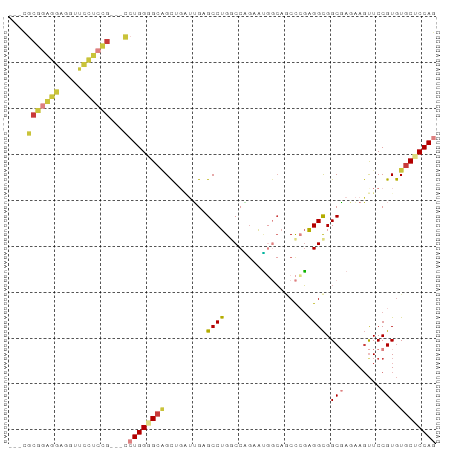
>dm3.chr3L 21656196 87 - 24543557 ---CGCGGAGGAGGUUCCUCCG---CCUGGAGCAGCUGAUUGCGCCUGGCCAGCAUGGCAGCCGGAGGCGGCGAGAAGUUCCGUGUGCUCCAG ---.(((((((.....))))))---)((((((((((.((...((((..(((.....))).(((...)))))))......)).)).)))))))) ( -44.30, z-score = -1.93, R) >droAna3.scaffold_13337 3980390 93 - 23293914 GGGUGCUGCUCCUGCUCCGGCAGUCUGUGGUGCUGUAGGAGGAGCCUGUCCGGAGCCACAACCGGAGGCGGCUAGCAGCUCCGUAUGCUCCAG .((.(((((((((.((.(((((.(....).))))).)).)))(((((.(((((........))))).).)))))))))).))........... ( -41.00, z-score = -0.13, R) >droWil1.scaffold_180698 10321159 84 + 11422946 -----CGAAUGGGAAUUGGCUGGCCCAUGG-GAGGC---AGGAGCCUGACCCGGAGUUGUACUCUGGGUGGCCAUAAUUUCCGUGUGCUCCAG -----...((((((((((..(((((.....-.((((---....)))).((((((((.....)))))))))))))))))))))))......... ( -36.80, z-score = -2.16, R) >droSec1.super_11 1576113 87 - 2888827 ---CGCGGAGGAAGUUCCUCCG---CCUGGGGCAGCUGAUUGCGCCUGGCCAGCAUGGCAUCCCGAGGCGGCGAGAAGUUCCGUGUGCUCCAG ---.(((((((.....))))))---)(((((((((((..((((((((.(((.....)))......)))).))))..)))......)))))))) ( -42.20, z-score = -1.67, R) >droSim1.chr3L 20999155 87 - 22553184 ---CGCGGAGGAAGUUCCUCCG---CCUGGGGCAGCUGAUUGCGCCUGGCCACCAUGGCAGCCCGAGGCGGCGAGAAGUUCCGUGUGCUCCAG ---.(((.(((((.((((.(((---(((.((((.((.....))((((((...))).))).)))).)))))).).))).)))).).)))..... ( -46.00, z-score = -2.67, R) >droEre2.scaffold_4784 21312719 87 - 25762168 ---CGCGGAGGAGGUUCCUCCG---CCUGGGGCAGCGGAUUGGGCCUGGCCGGAAUGGCAGCCCGAGGCGGCGAGAAGUUCCGUGUGCUCCAG ---.(((((((.....))))))---)(((((((((((((((((((...(((.....))).))))))....((.....))))))).)))))))) ( -47.50, z-score = -2.29, R) >droYak2.chr3L 5341880 84 + 24197627 ---CGCGGAGGAGGUUCCUCCG---CCUGGAGCAGC---UUGAGCCUGCCCAGAAUGGCAGCCCGAGGCGGCGAGAAGUUCCGUGUGCUCCAG ---.(((((((.....))))))---)((((((((.(---(((.((.((((......)))))).))))((((.........)))).)))))))) ( -43.00, z-score = -2.92, R) >consensus ___CGCGGAGGAGGUUCCUCCG___CCUGGGGCAGCUGAUUGAGCCUGGCCAGAAUGGCAGCCCGAGGCGGCGAGAAGUUCCGUGUGCUCCAG .....((((((.....))))))....((((((((.........((((..................)))).(((........))).)))))))) (-19.10 = -20.17 + 1.07)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:42:45 2011