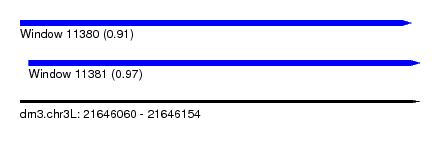
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 21,646,060 – 21,646,154 |
| Length | 94 |
| Max. P | 0.973932 |

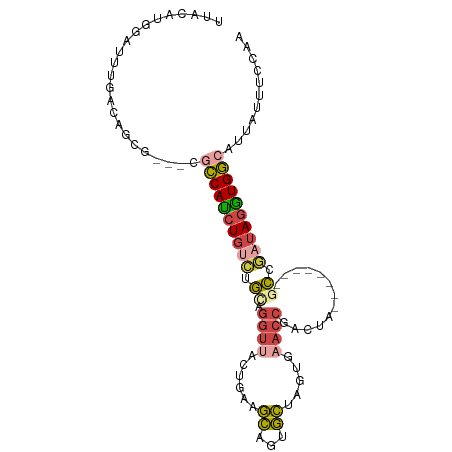
| Location | 21,646,060 – 21,646,152 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 67.68 |
| Shannon entropy | 0.58003 |
| G+C content | 0.46628 |
| Mean single sequence MFE | -28.60 |
| Consensus MFE | -11.80 |
| Energy contribution | -12.44 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.44 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.41 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.22 |
| SVM RNA-class probability | 0.912078 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 21646060 92 + 24543557 AGUUACGUGGAUUUGACAGCGUG-CGCCAUCUGUCUGCAGGUUACUGAAGCAGUGCUAGUGAACCGACUAU-------GCCGAUAGGUGGCGUUAUUUCC ........(((..((((.(....-)((((((((((.((((((((((..(((...))))))).))).....)-------)).))))))))))))))..))) ( -31.20, z-score = -1.58, R) >droAna3.scaffold_13337 3970899 99 + 23293914 AAUUGUAUUCCUUUAAAGACCUGAGGUCAUCUAGUUAUUACAUACUUUAAUUUAAUGAAGAUGCCUAUUAGAAUGCCAUUCAAAAAGUGAAAGUCUUCC- ...............(((((....(((..((((((.....(((.(((((......))))))))...))))))..))).((((.....)))).)))))..- ( -15.70, z-score = -0.81, R) >droEre2.scaffold_4784 21302628 88 + 25762168 AGCUGCAUGGAUUUGACAGCG-----CCAUCUGUCUGUAGGUUACUCAAGCAGUGCUGGUGAACCGGCUAU-------GCCGAUAGAUGGCACUAUUUCA .((((((......)).))))(-----(((((((((((.((....)))).(((..(((((....)))))..)-------)).))))))))))......... ( -37.70, z-score = -3.70, R) >droYak2.chr3L 5331090 91 - 24197627 AGAUACAUUUAUUUAACAGCGUG-CGCCACCUGGCCGUAGGUUACACAAGCAGAGCUGGUGUACCGGCUU--------UCCAAUAGGUGGCAUUAUUUCA .......................-.(((((((((((...))).......(.((((((((....)))))))--------).)..))))))))......... ( -26.80, z-score = -1.28, R) >droSec1.super_11 1565982 89 + 2888827 AGUUACAUGGAUUUGACAGCA---CGCCAUCUGUCUGCAGGUUACUGAAGCAGUGCCUGUGAACCGACUA--------GCCGAUAGGUGGCGUUAUUUCC ........(((..((.....(---(((((((((((.((((((.((((...)))))))))).....(....--------..)))))))))))))))..))) ( -30.10, z-score = -2.05, R) >droSim1.chr3L 20988750 89 + 22553184 AGUUACAUGGAUUUGACAGCA---CGCCAUCUGUCUGCAGGUUACUGAAGCAGUGCCUGUGAACCGACUA--------GCCGAUAGGUGGCGUUAUUUCC ........(((..((.....(---(((((((((((.((((((.((((...)))))))))).....(....--------..)))))))))))))))..))) ( -30.10, z-score = -2.05, R) >consensus AGUUACAUGGAUUUGACAGCG___CGCCAUCUGUCUGCAGGUUACUGAAGCAGUGCUAGUGAACCGACUA________GCCGAUAGGUGGCAUUAUUUCC .........................((((((((((.((.((((......((...)).....)))).............)).))))))))))......... (-11.80 = -12.44 + 0.64)

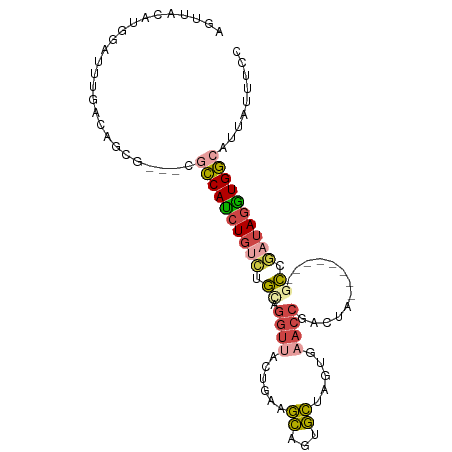
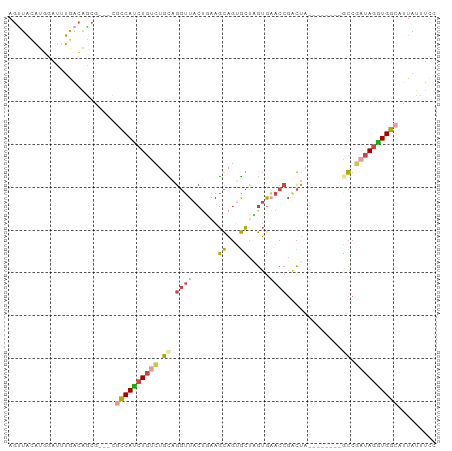
| Location | 21,646,062 – 21,646,154 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 67.33 |
| Shannon entropy | 0.58653 |
| G+C content | 0.45804 |
| Mean single sequence MFE | -29.20 |
| Consensus MFE | -11.80 |
| Energy contribution | -12.44 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.44 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.90 |
| SVM RNA-class probability | 0.973932 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 21646062 92 + 24543557 UUACGUGGAUUUGACAGCGUG-CGCCAUCUGUCUGCAGGUUACUGAAGCAGUGCUAGUGAACCGACUAU-------GCCGAUAGGUGGCGUUAUUUCCAA .....((((..((((.(....-)((((((((((.((((((((((..(((...))))))).))).....)-------)).))))))))))))))..)))). ( -33.80, z-score = -2.50, R) >droAna3.scaffold_13337 3970901 97 + 23293914 UUGUAUUCCUUUAAAGACCUGAGGUCAUCUAGUUAUUACAUACUUUAAUUUAAUGAAGAUGCCUAUUAGAAUGCCAUUCAAAAAGUGAAAGUCUUCC--- .............(((((....(((..((((((.....(((.(((((......))))))))...))))))..))).((((.....)))).)))))..--- ( -15.70, z-score = -0.85, R) >droEre2.scaffold_4784 21302630 88 + 25762168 CUGCAUGGAUUUGACAGCG-----CCAUCUGUCUGUAGGUUACUCAAGCAGUGCUGGUGAACCGGCUAU-------GCCGAUAGAUGGCACUAUUUCAAA .........(((((.((.(-----(((((((((((.((....)))).(((..(((((....)))))..)-------)).)))))))))).))...))))) ( -33.30, z-score = -2.91, R) >droYak2.chr3L 5331092 91 - 24197627 AUACAUUUAUUUAACAGCGUG-CGCCACCUGGCCGUAGGUUACACAAGCAGAGCUGGUGUACCGGCUU--------UCCAAUAGGUGGCAUUAUUUCAAA .....................-.(((((((((((...))).......(.((((((((....)))))))--------).)..))))))))........... ( -26.80, z-score = -1.38, R) >droSec1.super_11 1565984 89 + 2888827 UUACAUGGAUUUGACAGCA---CGCCAUCUGUCUGCAGGUUACUGAAGCAGUGCCUGUGAACCGACUA--------GCCGAUAGGUGGCGUUAUUUCCAA .....((((..((.....(---(((((((((((.((((((.((((...)))))))))).....(....--------..)))))))))))))))..)))). ( -32.80, z-score = -3.06, R) >droSim1.chr3L 20988752 89 + 22553184 UUACAUGGAUUUGACAGCA---CGCCAUCUGUCUGCAGGUUACUGAAGCAGUGCCUGUGAACCGACUA--------GCCGAUAGGUGGCGUUAUUUCCAA .....((((..((.....(---(((((((((((.((((((.((((...)))))))))).....(....--------..)))))))))))))))..)))). ( -32.80, z-score = -3.06, R) >consensus UUACAUGGAUUUGACAGCG___CGCCAUCUGUCUGCAGGUUACUGAAGCAGUGCUAGUGAACCGACUA________GCCGAUAGGUGGCAUUAUUUCCAA .......................((((((((((.((.((((......((...)).....)))).............)).))))))))))........... (-11.80 = -12.44 + 0.64)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:42:44 2011