| Sequence ID | dm3.chr2L |
|---|---|
| Location | 6,023,425 – 6,023,519 |
| Length | 94 |
| Max. P | 0.617382 |

| Location | 6,023,425 – 6,023,519 |
|---|---|
| Length | 94 |
| Sequences | 11 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 72.91 |
| Shannon entropy | 0.52382 |
| G+C content | 0.48726 |
| Mean single sequence MFE | -33.34 |
| Consensus MFE | -9.36 |
| Energy contribution | -9.00 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.36 |
| Mean z-score | -2.43 |
| Structure conservation index | 0.28 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.617382 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
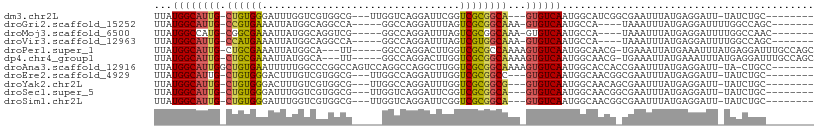
>dm3.chr2L 6023425 94 + 23011544 UUAUGGCAUUG-CUGUGGGAUUUGGUCGUGGCG---UUGGUCAGGAUUCGGUCGCGGCA---GUGUCAAUGGCAUCGGCGAAUUUAUGAGGAUU-UAUCUGC-------- ...((((((((-((((..(((..((((.((((.---...)))).))))..)))))))))---))))))...(((..(..((((((....)))))-)..))))-------- ( -32.30, z-score = -2.46, R) >droGri2.scaffold_15252 2902672 92 + 17193109 UUAUGGCAUUG-CCGUGAAAUUAUGGCAGGCCA-----GGCCAGGAUUUAGUCGCGGCAAA-GUGUCAAUGCCA----UAAAUUUAUGAGGAUUUUGGCCAGC------- ...((((.(((-(((((....))))))))))))-----(((((((((((.(.(((......-))).).....((----((....)))).)))))))))))...------- ( -34.70, z-score = -3.21, R) >droMoj3.scaffold_6500 13695512 92 + 32352404 UUAUGGCCAUG-CGGCGAAAUUAUGGCAGGUCG-----GGCCAGGAUUUAGUCGCGGCAAA-GUGUCAAUGCCA----UAAAUUUAUGAGGAUUUUGGCCAAC------- ...((((((.(-((((.(((((.((((......-----.)))).))))).)))))((((..-.......)))).----.................))))))..------- ( -32.40, z-score = -2.68, R) >droVir3.scaffold_12963 3326314 92 + 20206255 UUAUGGCAUUG-CCAUGAAAUUAUGGCAGGCCA-----GGCCAGGAUUUAGUCGUGGCAAA-GUGUCAAUGCCA----UAAAUUUAUGAGGAUUUUGGCCAGC------- ...((((.(((-(((((....))))))))))))-----(((((((((((.((..((((...-..))))..))((----((....)))).)))))))))))...------- ( -35.40, z-score = -3.55, R) >droPer1.super_1 10037414 100 - 10282868 UUAUGGCAUUG-CUGCGAAAUUAUGGCA---UU-----GGCCAGGACUUGGUCGCGCCAAAAGUGUCAAUGGCAACG-UGAAAUUAUGAAAUUUAUGAGGAUUUGCCAGC ...((((((((-(.((((.....((((.---..-----.))))........))))))....))))))).((((((((-((((.........)))))).....)))))).. ( -25.82, z-score = -0.01, R) >dp4.chr4_group1 5029791 100 - 5278887 UUAUGGCAUUG-CUGCGAAAUUAUGGCA---UU-----GGCCAGGACUUGGUCGCGGCAAAAGUGUCAAUGGCAACG-UGAAAUUAUGAAAUUUAUGAGGAUUUGCCAGC ...((((((((-((((((.....((((.---..-----.))))........))))))))...)))))).((((((((-((((.........)))))).....)))))).. ( -32.12, z-score = -2.27, R) >droAna3.scaffold_12916 11438394 101 + 16180835 UUAUGGCAUUGGCUGUGAAUUUUUGGCCCGGCCAGUCCAGGCCAGGCUUGGUCGCGGCAAAAGUGUCAAUGGCACCACCGAAUUUAUGAGGAUU-UA-CUGCC------- ...(((((((.(((((((......((((.((((......)))).))))...)))))))...)))))))..((((.....((((((....)))))-).-.))))------- ( -37.50, z-score = -1.53, R) >droEre2.scaffold_4929 15539776 94 - 26641161 UUAUGGCAUUG-CUGUGGGACUUUGUCGUGGCG---UUGGCCAGGAUUUGGUCGCGGCC---GUGUCAAUGGCAACGGCGAAUUUAUGAGGAUU-UAUCUGC-------- ((((((..(((-(((((.((((..(((.((((.---...)))).)))..)))).).(((---((....))))).)))))))..)))))).....-.......-------- ( -35.40, z-score = -2.51, R) >droYak2.chr2L 9154267 94 - 22324452 UUAUGGCAUUG-CUGUGGGACUUUGUCGUGGCG---UUGGCCAGGAUUUGGUCGCGGCG---GUGUCAAUGGCAACAGCGAAUUUAUGAGGAUU-UAUCUGC-------- ...((((((((-((((..((((..(((.((((.---...)))).)))..))))))))))---)))))).((....))..((((((....)))))-)......-------- ( -35.90, z-score = -3.16, R) >droSec1.super_5 4094396 94 + 5866729 UUAUGGCAUUG-CUGUGGGAUUUGGUCGUGGCG---UUGGUCAGGAUUCGGUCGCGGCA---GUGUCAAUGGCAACGGCGAAUUUAUGAGGAUU-UAUCUGC-------- ...((((((((-((((..(((..((((.((((.---...)))).))))..)))))))))---))))))..(....)(..((((((....)))))-)..)...-------- ( -32.60, z-score = -2.67, R) >droSim1.chr2L 5811862 94 + 22036055 UUAUGGCAUUG-CUGUGGGAUUUGGUCGUGGCG---UUGGUCAGGAUUCGGUCGCGGCA---GUGUCAAUGGCAACGGCGAAUUUAUGAGGAUU-UAUCUGC-------- ...((((((((-((((..(((..((((.((((.---...)))).))))..)))))))))---))))))..(....)(..((((((....)))))-)..)...-------- ( -32.60, z-score = -2.67, R) >consensus UUAUGGCAUUG_CUGUGAAAUUUUGGCAUGGCG_____GGCCAGGAUUUGGUCGCGGCAAA_GUGUCAAUGGCAACG_CGAAUUUAUGAGGAUU_UAUCUGC________ ...((((((((.((((((.................................))))))))...)))))).......................................... ( -9.36 = -9.00 + -0.36)
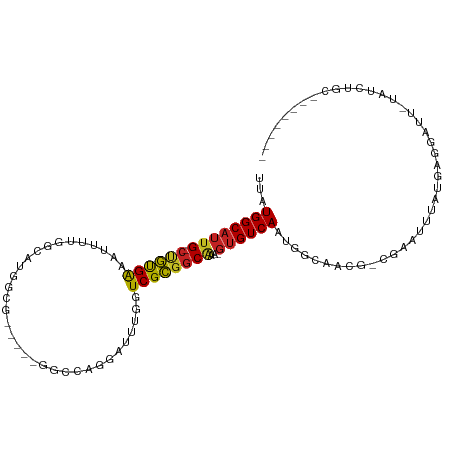
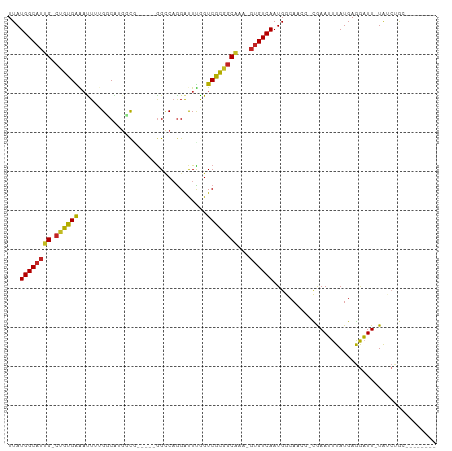
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:20:19 2011