
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 21,599,313 – 21,599,410 |
| Length | 97 |
| Max. P | 0.951594 |

| Location | 21,599,313 – 21,599,410 |
|---|---|
| Length | 97 |
| Sequences | 7 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 84.05 |
| Shannon entropy | 0.29610 |
| G+C content | 0.52165 |
| Mean single sequence MFE | -29.30 |
| Consensus MFE | -20.20 |
| Energy contribution | -21.39 |
| Covariance contribution | 1.19 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.69 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.45 |
| SVM RNA-class probability | 0.939031 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
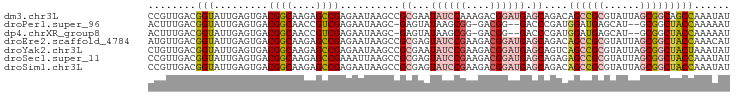
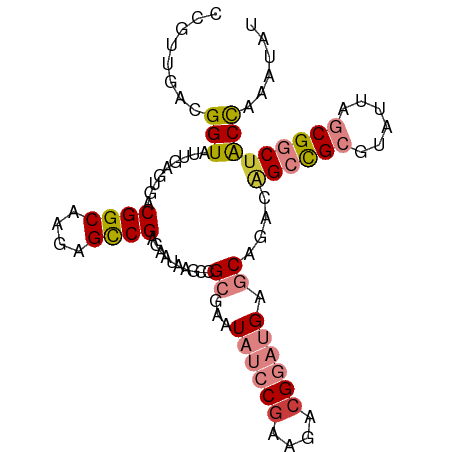
>dm3.chr3L 21599313 97 + 24543557 CCGUUGACGGUAUUGAGUGACGGCAAGAGCCGAGAAUAAGCCGCGAAUAUCCAAAGACGGAUGAGCAGACAGCCGCGUAUUAGCGGCAGCCAAAUAU ........(((.....((..((((....))))..........((...(((((......))))).))..)).(((((......))))).)))...... ( -28.60, z-score = -2.30, R) >droPer1.super_96 185493 91 - 202244 ACUUUGACGGUAUUGAGUGACGGCAACCGUCGAGAAUAAGC-GAGUAUAAGCGG-GACGG--GACCCGAUGGAUGAGCAU--GCGGCUACCAAAAAU ........(((((((.(((.(..((.(((((........((-........))((-(....--..)))))))).)).))))--.))).))))...... ( -23.00, z-score = -0.53, R) >dp4.chrXR_group8 451250 91 + 9212921 ACUUUGACGGUAUUGAGUGACGGCAACCGUCGAGAAUAAGC-GAGUAUAAGCGG-GACGG--GACCCGAUGGAUGAGCAU--GCGGCUACCAAAAAU ........(((((((.(((.(..((.(((((........((-........))((-(....--..)))))))).)).))))--.))).))))...... ( -23.00, z-score = -0.53, R) >droEre2.scaffold_4784 21254380 97 + 25762168 AUGUUGACGGUAUUGAGUGACGGCAAGAGCCGAGAAUAAGCCGCGAGUAUCCGAAGACGGAUGAGCAGACAGCCGCGUAUUAGCGGCUACCAAACAU (((((...(((.....((..((((....))))..........((...((((((....)))))).))..))((((((......))))))))).))))) ( -33.30, z-score = -3.60, R) >droYak2.chr3L 5285088 97 - 24197627 CUGUUGACGGUAUUGAGUGACGGCAAGAGCCGAGAAUAAGCCGCGAAUAUCCGAAGACGGAUGAGCAGUCAGCCGCGUAUUAGCGGCUACUAAAUAU .....((((((.(((..(..((((....)))).)..))))))((...((((((....)))))).)).)))((((((......))))))......... ( -32.50, z-score = -2.79, R) >droSec1.super_11 1519571 97 + 2888827 CCGUUGACGGUAUUGAGUGACGGCAAGAGCCGAAAUUAAGCCGCGAGUAUCCGAAGACGGAUGAGCAGAGAGCCGCGUAUUAGCGGCUACCAAAUAU .....(.((((.((((....((((....))))...)))))))))(..((((((....))))))..).(..((((((......))))))..)...... ( -32.90, z-score = -3.44, R) >droSim1.chr3L 20936163 97 + 22553184 CCGUUGACGGUAUUGAGUGACGGCAAGAGCCGAGAAUAAGCCGCGAGUAUCCGAAGACGGAUGAGCAGACAGCCGCGUAUUAGCGGCUACCAAAUAU ........(((.....((..((((....))))..........((...((((((....)))))).))..))((((((......)))))))))...... ( -31.80, z-score = -3.03, R) >consensus CCGUUGACGGUAUUGAGUGACGGCAAGAGCCGAGAAUAAGCCGCGAAUAUCCGAAGACGGAUGAGCAGACAGCCGCGUAUUAGCGGCUACCAAAUAU ........(((.........((((....))))..........((...((((((....)))))).))....((((((......)))))))))...... (-20.20 = -21.39 + 1.19)
| Location | 21,599,313 – 21,599,410 |
|---|---|
| Length | 97 |
| Sequences | 7 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 84.05 |
| Shannon entropy | 0.29610 |
| G+C content | 0.52165 |
| Mean single sequence MFE | -27.40 |
| Consensus MFE | -17.73 |
| Energy contribution | -19.14 |
| Covariance contribution | 1.41 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.53 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.57 |
| SVM RNA-class probability | 0.951594 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
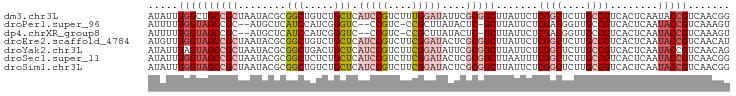
>dm3.chr3L 21599313 97 - 24543557 AUAUUUGGCUGCCGCUAAUACGCGGCUGUCUGCUCAUCCGUCUUUGGAUAUUCGCGGCUUAUUCUCGGCUCUUGCCGUCACUCAAUACCGUCAACGG .....((((.(((((......((((....))))..(((((....)))))....)))))........(((....))))))).......(((....))) ( -28.90, z-score = -2.48, R) >droPer1.super_96 185493 91 + 202244 AUUUUUGGUAGCCGC--AUGCUCAUCCAUCGGGUC--CCGUC-CCGCUUAUACUC-GCUUAUUCUCGACGGUUGCCGUCACUCAAUACCGUCAAAGU ......(((((((((--.............(((..--....)-))((........-))........).))))))))..................... ( -19.70, z-score = -0.85, R) >dp4.chrXR_group8 451250 91 - 9212921 AUUUUUGGUAGCCGC--AUGCUCAUCCAUCGGGUC--CCGUC-CCGCUUAUACUC-GCUUAUUCUCGACGGUUGCCGUCACUCAAUACCGUCAAAGU ......(((((((((--.............(((..--....)-))((........-))........).))))))))..................... ( -19.70, z-score = -0.85, R) >droEre2.scaffold_4784 21254380 97 - 25762168 AUGUUUGGUAGCCGCUAAUACGCGGCUGUCUGCUCAUCCGUCUUCGGAUACUCGCGGCUUAUUCUCGGCUCUUGCCGUCACUCAAUACCGUCAACAU (((((((((((((((......((((....))))..(((((....)))))....))))).......((((....))))........)))))..))))) ( -32.10, z-score = -3.64, R) >droYak2.chr3L 5285088 97 + 24197627 AUAUUUAGUAGCCGCUAAUACGCGGCUGACUGCUCAUCCGUCUUCGGAUAUUCGCGGCUUAUUCUCGGCUCUUGCCGUCACUCAAUACCGUCAACAG .....((((((((((......)))))).))))...(((((....)))))....((((........((((....))))..........))))...... ( -29.37, z-score = -3.06, R) >droSec1.super_11 1519571 97 - 2888827 AUAUUUGGUAGCCGCUAAUACGCGGCUCUCUGCUCAUCCGUCUUCGGAUACUCGCGGCUUAAUUUCGGCUCUUGCCGUCACUCAAUACCGUCAACGG .((((.(((((((((......((((....))))..(((((....)))))....))))))......((((....))))..))).))))(((....))) ( -31.00, z-score = -3.64, R) >droSim1.chr3L 20936163 97 - 22553184 AUAUUUGGUAGCCGCUAAUACGCGGCUGUCUGCUCAUCCGUCUUCGGAUACUCGCGGCUUAUUCUCGGCUCUUGCCGUCACUCAAUACCGUCAACGG .((((.(((((((((......((((....))))..(((((....)))))....))))))......((((....))))..))).))))(((....))) ( -31.00, z-score = -3.15, R) >consensus AUAUUUGGUAGCCGCUAAUACGCGGCUGUCUGCUCAUCCGUCUUCGGAUAUUCGCGGCUUAUUCUCGGCUCUUGCCGUCACUCAAUACCGUCAACGG .....((((((((((........(((.....))).(((((....)))))....))))).......((((....))))........)))))....... (-17.73 = -19.14 + 1.41)
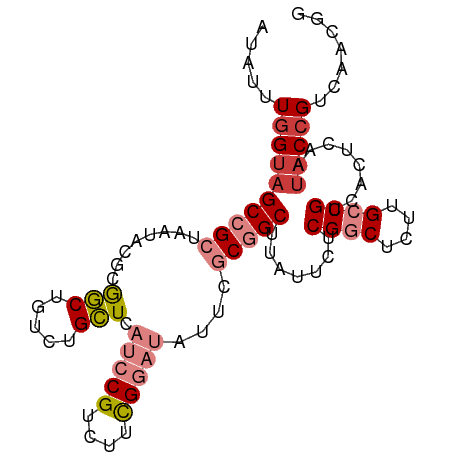
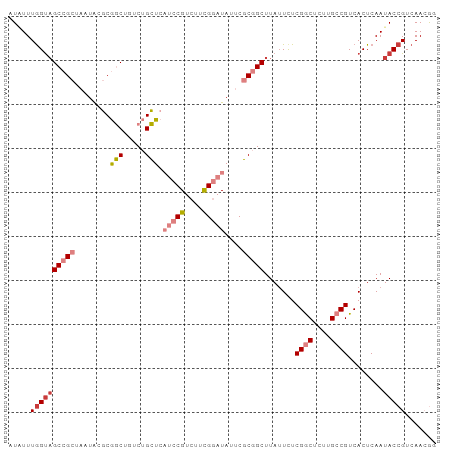
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:42:40 2011