| Sequence ID | dm3.chr3L |
|---|---|
| Location | 21,595,880 – 21,595,970 |
| Length | 90 |
| Max. P | 0.999671 |

| Location | 21,595,880 – 21,595,970 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 59.60 |
| Shannon entropy | 0.71398 |
| G+C content | 0.60889 |
| Mean single sequence MFE | -35.38 |
| Consensus MFE | -16.21 |
| Energy contribution | -16.49 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.53 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.55 |
| SVM RNA-class probability | 0.992612 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
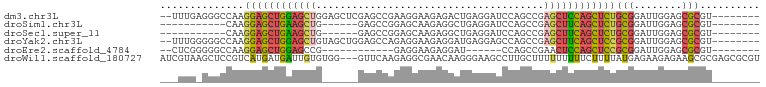
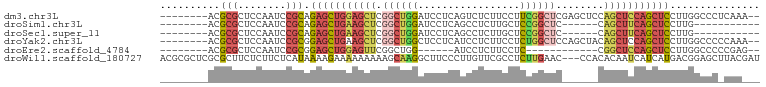
>dm3.chr3L 21595880 90 + 24543557 --UUUGAGGGCCAAGGAGCUGGAGCUGGAGCUCGAGCCGAAGGAAGAGACUGAGGAUCCAGCCGAGCUCCAGCUCUGCGGAUUGGAGCGCGU-------- --((..(...((..((((((((((((.(.(((.((.((..((.......))..)).)).)))).))))))))))))..)).)..))......-------- ( -40.90, z-score = -2.05, R) >droSim1.chr3L 20932535 75 + 22553184 -----------CAAGGAGCUGAAGCUG------GAGCCGGAGCAAGAGGCUGAGGAUCCAGCCGAGCUUCAGCUCUGCGGAUUGGAGCGCGU-------- -----------...((((((((((((.------..((....))....(((((......))))).))))))))))))(((........)))..-------- ( -34.50, z-score = -2.08, R) >droSec1.super_11 1516009 75 + 2888827 -----------CAAGGAGCUGAAGCUG------GAGCCGGAGCAAGAGGCUGAGGAUCCAGCCGAGCUUCAGCUCUGCGGAUUGGAGCGCGU-------- -----------...((((((((((((.------..((....))....(((((......))))).))))))))))))(((........)))..-------- ( -34.50, z-score = -2.08, R) >droYak2.chr3L 5281732 90 - 24197627 --UUUGGGGGCCAAGGAGCUGGAGCUGUAGCUGGAGCCAGAGGAAGAGGAUGAGGAGCCAGCCGAGCUUCAGCUCCGCGGAUUGGAGCGCGU-------- --.((((...))))((((((((((((...(((((..((...............))..)))))..))))))))))))(((........)))..-------- ( -39.46, z-score = -2.07, R) >droEre2.scaffold_4784 21250989 72 + 25762168 --CUCGGGGGCCAAGGAGCUGGAGCCG------------GAGGAAGAGGAU------CCAGCCGAACUCCAGCUCCGCGGAUUGGAGCGCGU-------- --(((.((..((..((((((((((.((------------(.(((......)------))..)))..))))))))))..)).)).))).....-------- ( -33.00, z-score = -1.37, R) >droWil1.scaffold_180727 1708519 97 - 2741493 AUCGUAAGCUCCGUCAUGAUGAUUGUGUGG---GUUCAAGAGGCGAACAAGGGAAGCCUUGCUUUUUUUUUCUUUUAUGAGAAGAGAAGCGCGAGCGCGU .......((.(((.((..(...)..)))))---((((..(((((...........)))))((((((((((((......))))))))))))..)))))).. ( -29.90, z-score = -1.07, R) >consensus ___UUG_GGGCCAAGGAGCUGAAGCUG______GAGCCGGAGGAAGAGGAUGAGGAUCCAGCCGAGCUUCAGCUCUGCGGAUUGGAGCGCGU________ ..............((((((((((((......................................))))))))))))(((........))).......... (-16.21 = -16.49 + 0.28)
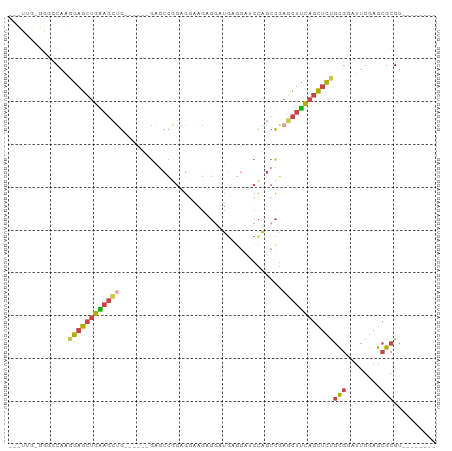
| Location | 21,595,880 – 21,595,970 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 59.60 |
| Shannon entropy | 0.71398 |
| G+C content | 0.60889 |
| Mean single sequence MFE | -31.09 |
| Consensus MFE | -15.93 |
| Energy contribution | -17.29 |
| Covariance contribution | 1.37 |
| Combinations/Pair | 1.52 |
| Mean z-score | -2.56 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 4.17 |
| SVM RNA-class probability | 0.999671 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
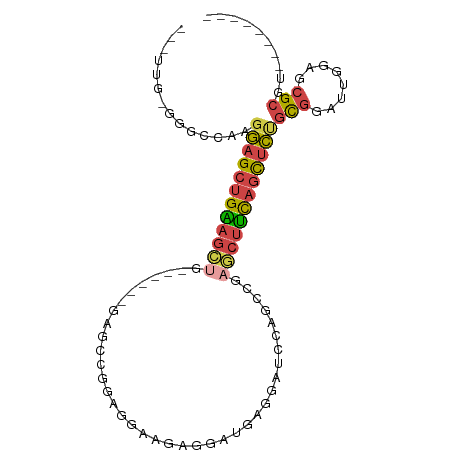
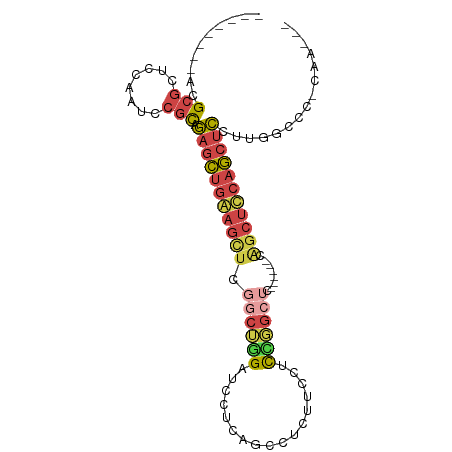
>dm3.chr3L 21595880 90 - 24543557 --------ACGCGCUCCAAUCCGCAGAGCUGGAGCUCGGCUGGAUCCUCAGUCUCUUCCUUCGGCUCGAGCUCCAGCUCCAGCUCCUUGGCCCUCAAA-- --------..(.(..((((...((.(((((((((((((((((((...............)))))).)))))))))))))..))...))))..).)...-- ( -38.66, z-score = -3.21, R) >droSim1.chr3L 20932535 75 - 22553184 --------ACGCGCUCCAAUCCGCAGAGCUGAAGCUCGGCUGGAUCCUCAGCCUCUUGCUCCGGCUC------CAGCUUCAGCUCCUUG----------- --------..(((........))).(((((((((((.((((((....))))))....((....))..------.)))))))))))....----------- ( -31.80, z-score = -3.12, R) >droSec1.super_11 1516009 75 - 2888827 --------ACGCGCUCCAAUCCGCAGAGCUGAAGCUCGGCUGGAUCCUCAGCCUCUUGCUCCGGCUC------CAGCUUCAGCUCCUUG----------- --------..(((........))).(((((((((((.((((((....))))))....((....))..------.)))))))))))....----------- ( -31.80, z-score = -3.12, R) >droYak2.chr3L 5281732 90 + 24197627 --------ACGCGCUCCAAUCCGCGGAGCUGAAGCUCGGCUGGCUCCUCAUCCUCUUCCUCUGGCUCCAGCUACAGCUCCAGCUCCUUGGCCCCCAAA-- --------..(((........)))(((((((.((((.((((((..((...............))..))))))..)))).)))))))((((...)))).-- ( -33.86, z-score = -3.03, R) >droEre2.scaffold_4784 21250989 72 - 25762168 --------ACGCGCUCCAAUCCGCGGAGCUGGAGUUCGGCUGG------AUCCUCUUCCUC------------CGGCUCCAGCUCCUUGGCCCCCGAG-- --------.....(((....(((.(((((((((((.(((..((------(......))).)------------))))))))))))).))).....)))-- ( -34.40, z-score = -3.11, R) >droWil1.scaffold_180727 1708519 97 + 2741493 ACGCGCUCGCGCUUCUCUUCUCAUAAAAGAAAAAAAAAGCAAGGCUUCCCUUGUUCGCCUCUUGAAC---CCACACAAUCAUCAUGACGGAGCUUACGAU ..((....))(((((((((((......))))......(((((((....)))))))............---...............)).)))))....... ( -16.00, z-score = 0.19, R) >consensus ________ACGCGCUCCAAUCCGCAGAGCUGAAGCUCGGCUGGAUCCUCAGCCUCUUCCUCCGGCUC______CAGCUCCAGCUCCUUGGCCC_CAA___ ..........(((........))).(((((((((((.(((((((...............)))))))........)))))))))))............... (-15.93 = -17.29 + 1.37)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:42:38 2011