| Sequence ID | dm3.chr3L |
|---|---|
| Location | 21,594,778 – 21,594,948 |
| Length | 170 |
| Max. P | 0.988782 |

| Location | 21,594,778 – 21,594,948 |
|---|---|
| Length | 170 |
| Sequences | 5 |
| Columns | 170 |
| Reading direction | forward |
| Mean pairwise identity | 93.38 |
| Shannon entropy | 0.11278 |
| G+C content | 0.45689 |
| Mean single sequence MFE | -52.88 |
| Consensus MFE | -47.36 |
| Energy contribution | -47.80 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.90 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.82 |
| SVM RNA-class probability | 0.969799 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
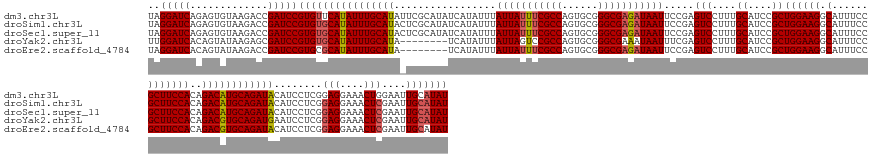
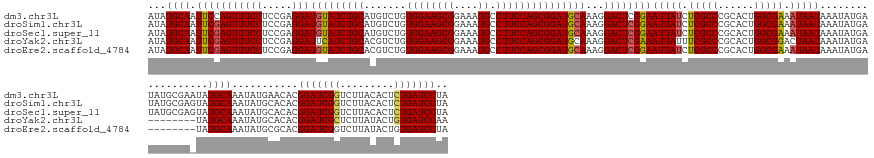
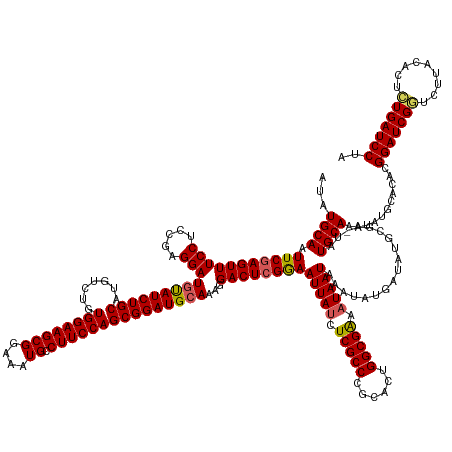
>dm3.chr3L 21594778 170 + 24543557 UAGGAUCAGAGUGUAAGACCGAUCCGUGUUCAUAUUUGCAUAUUCGCAUAUCAUAUUUAUUAUUUCGCCAGUGCGGGCGAGAUAAUUCCGAGUCCUUUGCAUCCGCUGGAAGGCAUUUCCGCUUCCACAGACAUGCAGAUACAUCCUCGGAGGAAACUGGAAUUGCAUAU ..(((((.(.((.....))))))))...........((((.((((.((..........(((((((((((......)))))))))))((((((...(((((((...((((.((((......)))).).)))..)))))))......))))))(....)))))))))))... ( -49.50, z-score = -1.39, R) >droSim1.chr3L 20931419 170 + 22553184 UAGGAUCAGAGUGUAAGACCGAUCCGUGUGCAUAUUUGCAUACUCGCAUAUCAUAUUUAUUAUUUCGCCAGUGCGGGCGAGAUAAUUCCGAGUCCUUUGCAUCCGCUGGAAGGCAUUUCCGCUUCCACAGACAUGCAGAUACAUCCUCGGAGGAAACUCGAAUUGCAUAU ..(((((.(.((.....))))))))(((((((....)))))))..(((..((......(((((((((((......)))))))))))..((((...(((((((...((((.((((......)))).).)))..)))))))......))))(((....)))))..))).... ( -56.90, z-score = -3.18, R) >droSec1.super_11 1514940 170 + 2888827 UAGGAUCAGAGUGUAAGACCGAUCCGUGUGCAUAUUUGCAUACUCGCAUAUCAUAUUUAUUAUUUCGCCAGUGCGGGCGAGAUAAUUCCGAGUCCUUUGCAUCCGCUGGAAGGCAUUUCCGCUUCCACAGACAUGCAGAUACAUCCUCGGAGGAAACUCGAAUUGCAUAU ..(((((.(.((.....))))))))(((((((....)))))))..(((..((......(((((((((((......)))))))))))..((((...(((((((...((((.((((......)))).).)))..)))))))......))))(((....)))))..))).... ( -56.90, z-score = -3.18, R) >droYak2.chr3L 5280571 162 - 24197627 UUGGAUCACAGUAUAAGAGCGAUCCGUGUGCAUAUUUGCAUA--------UCAUAUUUAUUAGUCCGCCAGUGCGGGCGAAAUAAUUUCGAGUCCUUUGCAUCCGCUGGAAGGCAUUUCCGCUUCCACAGACGUGCAGAUGAAUCCUCGGAGGAAACUCGAAUUGCAUAU ..(((((.(.........).)))))(((((((.((((.....--------...(((((....((((((....)))))).)))))....((((((.(((((((...((((.((((......)))).).)))..))))))).))...))))(((....)))))))))))))) ( -50.50, z-score = -1.81, R) >droEre2.scaffold_4784 21249859 162 + 25762168 UAGGAUCACAGUAUAAGACCGAUCCGUGCGCAUAUUUGCAUA--------UCAUAUUUAUUAUUUCGCCAGUGCGGGCGAGAUAAUUCCGAGUCCUUUGCAUCCGCUGGAAGGCAUUUCCGCUUCCACAGACGUGCAGAUACAUCCUCGGAGGAAACUCGAAUUGCAUAU ..(((((.............)))))(((((((....)))...--------((......(((((((((((......)))))))))))..((((...(((((((...((((.((((......)))).).)))..)))))))......))))(((....)))))...)))).. ( -50.62, z-score = -2.38, R) >consensus UAGGAUCAGAGUGUAAGACCGAUCCGUGUGCAUAUUUGCAUA_UCGCAUAUCAUAUUUAUUAUUUCGCCAGUGCGGGCGAGAUAAUUCCGAGUCCUUUGCAUCCGCUGGAAGGCAUUUCCGCUUCCACAGACAUGCAGAUACAUCCUCGGAGGAAACUCGAAUUGCAUAU ..(((((.............)))))((((((((((((((((.................(((((((((((......))))))))))).....(((....((....))((((((.(......)))))))..))))))))))))........(((....)))....))))))) (-47.36 = -47.80 + 0.44)
| Location | 21,594,778 – 21,594,948 |
|---|---|
| Length | 170 |
| Sequences | 5 |
| Columns | 170 |
| Reading direction | reverse |
| Mean pairwise identity | 93.38 |
| Shannon entropy | 0.11278 |
| G+C content | 0.45689 |
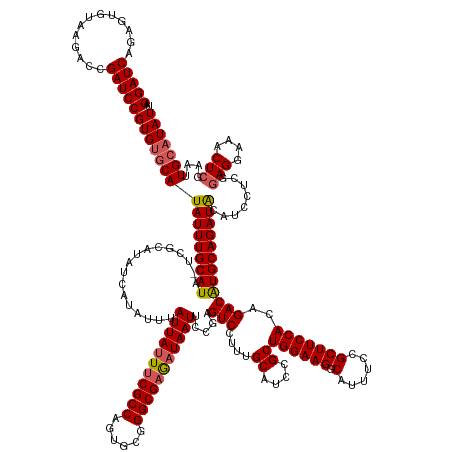
| Mean single sequence MFE | -54.67 |
| Consensus MFE | -51.03 |
| Energy contribution | -51.11 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.62 |
| Structure conservation index | 0.93 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.34 |
| SVM RNA-class probability | 0.988782 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 21594778 170 - 24543557 AUAUGCAAUUCCAGUUUCCUCCGAGGAUGUAUCUGCAUGUCUGUGGAAGCGGAAAUGCCUUCCAGCGGAUGCAAAGGACUCGGAAUUAUCUCGCCCGCACUGGCGAAAUAAUAAAUAUGAUAUGCGAAUAUGCAAAUAUGAACACGGAUCGGUCUUACACUCUGAUCCUA ...((((((((((...((.((((((..(((((((((.......((((((((....)).))))))))))))))).....))))))(((((.(((((......))))).)))))......))..)).)))).))))...........(((((((.........))))))).. ( -55.11, z-score = -2.96, R) >droSim1.chr3L 20931419 170 - 22553184 AUAUGCAAUUCGAGUUUCCUCCGAGGAUGUAUCUGCAUGUCUGUGGAAGCGGAAAUGCCUUCCAGCGGAUGCAAAGGACUCGGAAUUAUCUCGCCCGCACUGGCGAAAUAAUAAAUAUGAUAUGCGAGUAUGCAAAUAUGCACACGGAUCGGUCUUACACUCUGAUCCUA ...((((.(((((((((((.....)))(((((((((.......((((((((....)).)))))))))))))))...))))))))(((((.(((((......))))).)))))..........)))).((.((((....)))).))(((((((.........))))))).. ( -57.51, z-score = -2.90, R) >droSec1.super_11 1514940 170 - 2888827 AUAUGCAAUUCGAGUUUCCUCCGAGGAUGUAUCUGCAUGUCUGUGGAAGCGGAAAUGCCUUCCAGCGGAUGCAAAGGACUCGGAAUUAUCUCGCCCGCACUGGCGAAAUAAUAAAUAUGAUAUGCGAGUAUGCAAAUAUGCACACGGAUCGGUCUUACACUCUGAUCCUA ...((((.(((((((((((.....)))(((((((((.......((((((((....)).)))))))))))))))...))))))))(((((.(((((......))))).)))))..........)))).((.((((....)))).))(((((((.........))))))).. ( -57.51, z-score = -2.90, R) >droYak2.chr3L 5280571 162 + 24197627 AUAUGCAAUUCGAGUUUCCUCCGAGGAUUCAUCUGCACGUCUGUGGAAGCGGAAAUGCCUUCCAGCGGAUGCAAAGGACUCGAAAUUAUUUCGCCCGCACUGGCGGACUAAUAAAUAUGA--------UAUGCAAAUAUGCACACGGAUCGCUCUUAUACUGUGAUCCAA ...((((.(((((((((((.....))).....((...((((((((((((((....)).)))))).))))))...))))))))))((((.((((((......)))))).))))........--------..))))...........(((((((.........))))))).. ( -52.20, z-score = -2.67, R) >droEre2.scaffold_4784 21249859 162 - 25762168 AUAUGCAAUUCGAGUUUCCUCCGAGGAUGUAUCUGCACGUCUGUGGAAGCGGAAAUGCCUUCCAGCGGAUGCAAAGGACUCGGAAUUAUCUCGCCCGCACUGGCGAAAUAAUAAAUAUGA--------UAUGCAAAUAUGCGCACGGAUCGGUCUUAUACUGUGAUCCUA ...(((..(((((((((((.....)))(((((((((((....))(((((((....)).))))).)))))))))...))))))))(((((.(((((......))))).)))))........--------...(((....)))))).((((((((.....)))..))))).. ( -51.00, z-score = -1.68, R) >consensus AUAUGCAAUUCGAGUUUCCUCCGAGGAUGUAUCUGCAUGUCUGUGGAAGCGGAAAUGCCUUCCAGCGGAUGCAAAGGACUCGGAAUUAUCUCGCCCGCACUGGCGAAAUAAUAAAUAUGAUAUGCGA_UAUGCAAAUAUGCACACGGAUCGGUCUUACACUCUGAUCCUA ...((((.(((((((((((.....)))(((((((((.......((((((((....)).)))))))))))))))...))))))))(((((.(((((......))))).)))))..................))))...........(((((((.........))))))).. (-51.03 = -51.11 + 0.08)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:42:37 2011