| Sequence ID | dm3.chr3L |
|---|---|
| Location | 21,593,498 – 21,593,598 |
| Length | 100 |
| Max. P | 0.977007 |

| Location | 21,593,498 – 21,593,598 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 59.03 |
| Shannon entropy | 0.77242 |
| G+C content | 0.36628 |
| Mean single sequence MFE | -17.46 |
| Consensus MFE | -6.99 |
| Energy contribution | -7.60 |
| Covariance contribution | 0.61 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.96 |
| SVM RNA-class probability | 0.977007 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
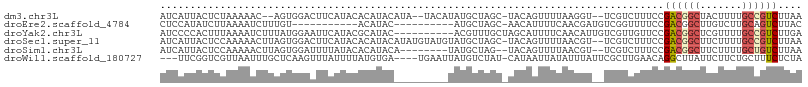
>dm3.chr3L 21593498 100 - 24543557 AUCAUUACUCUAAAAAC--AGUGGACUUCAUACACAUACAUA--UACAUAUGCUAGC-UACAGUUUUAAGGU--UCGUCUUUCCGACGGCUACUUUUGCCGUCUUAA ........(((.(((((--.((((.((.((((..........--....))))..)))-))).))))).))).--..........((((((.......)))))).... ( -19.84, z-score = -2.44, R) >droEre2.scaffold_4784 21248584 85 - 25762168 CUCCAUAUCUUAAAAUCUUUGU-----------ACAUAC----------AUGCUAGC-AACAUUUUCAACGAUGUCGGUUUUCCGACGGCUUGUCUUGCAGUCUUAC ..................((((-----------(...((----------(.(((...-...(((......)))(((((....)))))))).)))..)))))...... ( -12.90, z-score = -0.43, R) >droYak2.chr3L 5279284 97 + 24197627 AUCCCCACUUUAAAAUCUUUAUGGAAUUCAUACGCAUAC----------ACGUUUGCUAGCAUUUUCAACAUUGUCGUUGUUCCGACGGCUCGUUUUGCCGUCUUGA ......................((((.......(((...----------.....))).........((((......))))))))((((((.......)))))).... ( -19.80, z-score = -1.61, R) >droSec1.super_11 1513687 104 - 2888827 AUCAUUACUCCAAAAACUUAGUGGACUUCAUACACAUACAUAUGUAUGUAUGCUAGC-UACAGUUUUAACGU--UCGUCUUUCCGACGGCUUCUUUUGCCGUCUUAA ............((((((..((((.((.((((((..(((....)))))))))..)))-))))))))).....--..........((((((.......)))))).... ( -25.10, z-score = -3.86, R) >droSim1.chr3L 20930165 95 - 22553184 AUCAUUACUCCAAAAACUUAGUGGAUUUUAUACACAUACA--------UAUGCUAG--UACAGUUUUAACGU--UCGUCUUUCCGACGGCUUCUUUUGCUGUCUUAA ............((((((..(((.........))).(((.--------.......)--)).)))))).....--..........((((((.......)))))).... ( -13.80, z-score = -0.68, R) >droWil1.scaffold_180727 1706140 99 + 2741493 ---UUCGGUCGUUAAUUUGCUCAAGUUUAUUUUAUGUGA----UGAAUUAUGUCUAU-CAUAAUUAUAUUUAUUCGCUUGAACAGGCUUAUUCUUCUGCUUUCUCUA ---..(((....(((((((.((((((..((...((((((----(....((((.....-)))))))))))..))..)))))).)))).))).....)))......... ( -13.30, z-score = -0.47, R) >consensus AUCAUUACUCUAAAAACUUAGUGGACUUCAUACACAUAC_________UAUGCUAGC_UACAGUUUUAACGU__UCGUCUUUCCGACGGCUUCUUUUGCCGUCUUAA ....................................................................................((((((.......)))))).... ( -6.99 = -7.60 + 0.61)
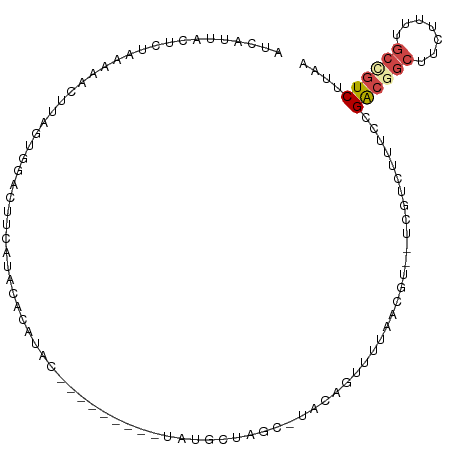

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:42:35 2011