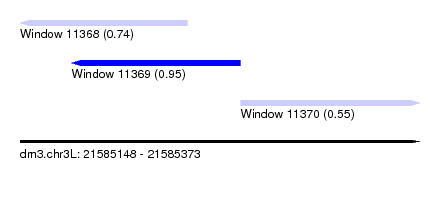
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 21,585,148 – 21,585,373 |
| Length | 225 |
| Max. P | 0.952381 |

| Location | 21,585,148 – 21,585,242 |
|---|---|
| Length | 94 |
| Sequences | 8 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 74.57 |
| Shannon entropy | 0.45226 |
| G+C content | 0.34131 |
| Mean single sequence MFE | -15.41 |
| Consensus MFE | -10.91 |
| Energy contribution | -11.24 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.03 |
| Structure conservation index | 0.71 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.744965 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 21585148 94 - 24543557 -----------UUCAGCACGAAAACGUGUUGAUACGAGAUAUUUCAACAUGUUCUGAAGCCAAUACGAU------UCCAAACCCAUGCCAUAAUUAAGCAUGUCUGCUAAU -----------...((((.((.((((((((((...........))))))))))................------........(((((.........)))))))))))... ( -18.90, z-score = -2.02, R) >droSim1.chr3L 20921442 94 - 22553184 -----------UUCAGCACGAAAACGUGUUGAUACGAGAUAUUUCAACAUGUUCUGAAGCCAAUACCAU------UCCAAAACCAUGCCAUAAUUAAGCAUGUUUGCUAAU -----------...((((....((((((((((...........))))))))))................------........(((((.........)))))..))))... ( -17.90, z-score = -1.95, R) >droSec1.super_11 1505273 94 - 2888827 -----------UUCAGCACGAAAACGUGUUGAUACGAGAUAUUUCAACAUGUUCUGAAGCCAAUACCAU------UCCAAAACCAUGCCAUAAUUAAGCAUGUUUGCUAAU -----------...((((....((((((((((...........))))))))))................------........(((((.........)))))..))))... ( -17.90, z-score = -1.95, R) >droYak2.chr3L 5270689 94 + 24197627 -----------UUCAGCACGAAAACGUGUUGAUAUGAGAUAUUUCAACAUGUUCUAGAGCCAAUACCAU------UCCAAAACCAUGCCAUAAUUAAGCAUGUUUGCUAAU -----------...(((((...((((((((((...........))))))))))...)............------........(((((.........)))))..))))... ( -18.00, z-score = -2.02, R) >droEre2.scaffold_4784 21239854 100 - 25762168 -----------UUCAUCACGAAGACGUGUUGAUAUGAGAUAUUUCAACAUGUUCUAAAGCCAAUACCAUGCCCAAUACCAAACCAUGCCAUAAUUAAGCAUGUUUGCUAAU -----------...........((((((((((...........))))))))))......................((.(((((.((((.........))))))))).)).. ( -16.80, z-score = -1.38, R) >droAna3.scaffold_13337 6239804 107 + 23293914 AAAUCACUGAAACCCCGGCCCCGACGUGUUGAUAUGGAAUAUUUCAACAUAUCUCGAAAAUAUCCAAAA----AACCAAUACCAAAGCAAUAAUUAAGCAUGUUUGCUAAU ................(((...((((((((((((((...........))))))..((.....)).....----.......................)))))))).)))... ( -13.20, z-score = -0.25, R) >dp4.chrXR_group8 433369 95 - 9212921 -----------UUUGAUACAAAGACUUGUUGAUUUGAAAUAUUUCAGCAUGUUCGGGGAACCUUAGCAA-----AUAAAAACAAAAACAAUAAUUAAGCAUGUUCACUAAU -----------..(((.(((..(((.((((((...........)))))).))).((....)).......-----..........................))))))..... ( -11.70, z-score = 0.24, R) >droPer1.super_96 160538 95 + 202244 -----------UUUGAUACAAAGACUUGUUGAUUUGAAAUAUUUCAGCAUGUUCGGAGAACCUUAGCAA-----AUAAAAACAAAAACAAUAAUUAAGCAUGUUCACUAAU -----------..(((.(((..(((.((((((...........)))))).))).((....)).......-----..........................))))))..... ( -8.90, z-score = 1.10, R) >consensus ___________UUCAGCACGAAAACGUGUUGAUAUGAGAUAUUUCAACAUGUUCUGAAGCCAAUACCAU______UCCAAAACCAUGCCAUAAUUAAGCAUGUUUGCUAAU ......................((((((((((...........))))))))))...........................................((((....))))... (-10.91 = -11.24 + 0.33)

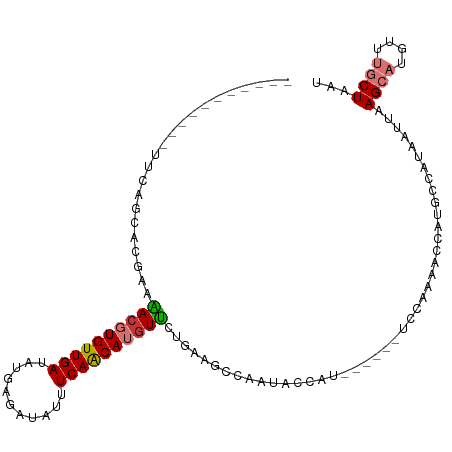
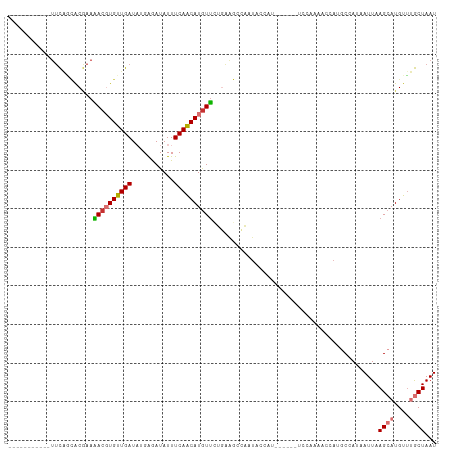
| Location | 21,585,177 – 21,585,272 |
|---|---|
| Length | 95 |
| Sequences | 8 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 65.18 |
| Shannon entropy | 0.64771 |
| G+C content | 0.35987 |
| Mean single sequence MFE | -15.11 |
| Consensus MFE | -9.56 |
| Energy contribution | -9.39 |
| Covariance contribution | -0.17 |
| Combinations/Pair | 1.30 |
| Mean z-score | -0.87 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.58 |
| SVM RNA-class probability | 0.952381 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
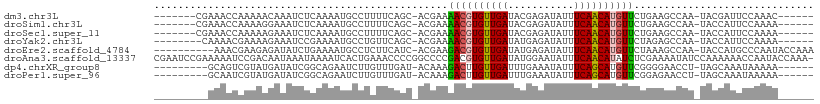
>dm3.chr3L 21585177 95 - 24543557 -------CGAAACCAAAAACAAAUCUCAAAAUGCCUUUUCAGC-ACGAAAACGUGUUGAUACGAGAUAUUUCAACAUGUUCUGAAGCCAA-UACGAUUCCAAAC------ -------((.............((((((((.....)))(((((-(((....))))))))...))))).(((((........)))))....-..)).........------ ( -14.30, z-score = -1.16, R) >droSim1.chr3L 20921471 95 - 22553184 -------CGAAACCAAAAGGAAAUCUCAAAAUGCCUUUUCAGC-ACGAAAACGUGUUGAUACGAGAUAUUUCAACAUGUUCUGAAGCCAA-UACCAUUCCAAAA------ -------...........((((((((((((.....)))(((((-(((....))))))))...))))).(((((........)))))....-.....))))....------ ( -18.30, z-score = -2.19, R) >droSec1.super_11 1505302 95 - 2888827 -------CGAAACCAAAAAGAAAUCUCAAAAUGCCUUUUCAGC-ACGAAAACGUGUUGAUACGAGAUAUUUCAACAUGUUCUGAAGCCAA-UACCAUUCCAAAA------ -------...........((((......(((((.(((.(((((-(((....))))))))...))).))))).......))))........-.............------ ( -14.92, z-score = -1.64, R) >droYak2.chr3L 5270718 94 + 24197627 --------CAAAACGAAAAGAAAUCCGAAAAUGCCUGUUCAGC-ACGAAAACGUGUUGAUAUGAGAUAUUUCAACAUGUUCUAGAGCCAA-UACCAUUCCAAAA------ --------..........((((....((((...((...(((((-(((....))))))))...).)...))))......))))........-.............------ ( -13.40, z-score = -0.79, R) >droEre2.scaffold_4784 21239883 98 - 25762168 ----------AAACGAAGAGAUAUCUGAAAAUGCCUCUUCAUC-ACGAAGACGUGUUGAUAUGAGAUAUUUCAACAUGUUCUAAAGCCAA-UACCAUGCCCAAUACCAAA ----------.......(((((((((.(.(((((.(((((...-..))))).)))))....).)))))))))..((((............-...))))............ ( -18.36, z-score = -2.34, R) >droAna3.scaffold_13337 6239829 109 + 23293914 CGAAUCCGAAAAAUCCGACAAUAAAUAAAAUCACUGAAACCCCGGCCCCGACGUGUUGAUAUGGAAUAUUUCAACAUAUCUCGAAAAUAUCCAAAAAACCAAUACCAAA- ......(((........................(((......)))....((..((((((...........))))))..)))))..........................- ( -10.50, z-score = 0.47, R) >dp4.chrXR_group8 433399 93 - 9212921 ---------GCAGUCGUAUGAUAUCGGCAGAAUCUUGUUUGAU-ACAAAGACUUGUUGAUUUGAAAUAUUUCAGCAUGUUCGGGGAACCU-UAGCAAAUAAAAA------ ---------...((((........))))......(((((((..-.....(((.((((((...........)))))).))).((....)).-...)))))))...------ ( -18.10, z-score = -0.38, R) >droPer1.super_96 160568 93 + 202244 ---------GCAAUCGUAUGAUAUCGGCAGAAUCUUGUUUGAU-ACAAAGACUUGUUGAUUUGAAAUAUUUCAGCAUGUUCGGAGAACCU-UAGCAAAUAAAAA------ ---------((.......((.(((((((((....))).)))))-)))..(((.((((((...........)))))).)))..........-..)).........------ ( -13.00, z-score = 1.07, R) >consensus _________AAAACAAAAAGAAAUCUGAAAAUGCCUGUUCAGC_ACGAAAACGUGUUGAUAUGAGAUAUUUCAACAUGUUCUGAAGCCAA_UACCAUUCCAAAA______ .................................................((((((((((...........)))))))))).............................. ( -9.56 = -9.39 + -0.17)
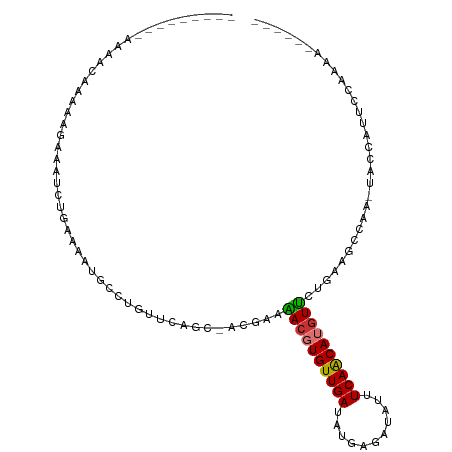

| Location | 21,585,272 – 21,585,373 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 71.39 |
| Shannon entropy | 0.54588 |
| G+C content | 0.41617 |
| Mean single sequence MFE | -21.65 |
| Consensus MFE | -10.75 |
| Energy contribution | -10.57 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.58 |
| Mean z-score | -1.09 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.553481 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
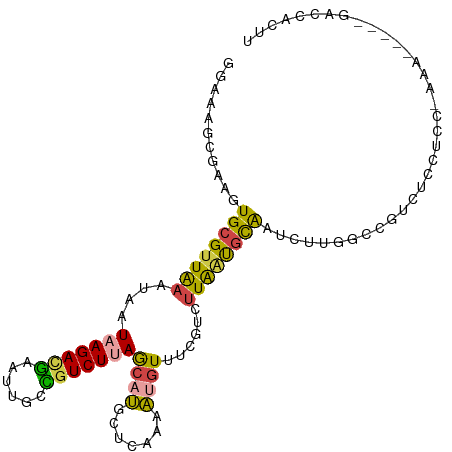
>dm3.chr3L 21585272 101 + 24543557 AGAAUGCGAAGUGCGUUAACGAAUAAGACGAAUUGUUGUCUUAGCAUGCUCAAAAUGUUUCGUCUUAAUGCAAUCUUGGCCAUCUCCUCC-GAA-----GACCACUU (((..((.(((((((((((((((((((((((....))))))))((((.......)))))))))..)))))))..))).))..))).....-...-----........ ( -23.80, z-score = -1.44, R) >droAna3.scaffold_13337 6239938 104 - 23293914 GGACAACACGUUGCGUUGAAUAAUAAGAUAAAUUACUGUCUUAACUGUCUCUCGUCAUGUAGUAGUGGC-UGGUUAAAAGUGUUUCUUUUUGCAAAA--GACUACUU .....((((((.(((..((....(((((((......)))))))....))...))).)))).))((((((-(.((.(((((.....))))).))...)--).))))). ( -21.50, z-score = -0.66, R) >droEre2.scaffold_4784 21239981 91 + 25762168 ----------GUGCGCUAAAUAAUAAGACGCAUUGCCGUCUCAGCCUGCUUUAAAUGUUUCUUUUUAAUGCAAUCAUGGCCGUCUCCUCCAAA------GACUACUU ----------(((((((........)).))))).(((((....(..(((.(((((........))))).)))..)))))).((((.......)------)))..... ( -16.30, z-score = -0.93, R) >droYak2.chr3L 5270812 107 - 24197627 GGAAAGCGAAGUGCGUUAAAUAAUAAGACGAAUUGCCAUCUCAGCAUGCUUCAAAUGUUUCUUUUUAAUGCAAUCUUGGCCGUCUCCUCCAACUACGCCAACUACUU .........(((((((.........(((((....((((.....((((....................)))).....))))))))).........))))..))).... ( -18.22, z-score = 0.01, R) >droSec1.super_11 1505397 102 + 2888827 GGAAUGCGAAGUGCGUUAACUAAUAAGACGAAUUGCCGUCUUAGCAUGCUCAAAAUGUUUCGUCUUAAUGCAAUCUUGGCCGUCUCCUCCCAAA-----GACCACUU (((..((.(((((((((((....(((((((......)))))))((((.......))))......))))))))..))).))....))).......-----........ ( -23.30, z-score = -1.51, R) >droSim1.chr3L 20921566 101 + 22553184 GGAACGCGAAGUGCGUUAACUAAUAAGACGAAUUGCCGUCUUAGCAUGCUCAAAAUGUUUCGUCUUAAUGCAAUCUUGGCCGUCUCCUCC-AGA-----GACCACUU .....((.(((((((((((....(((((((......)))))))((((.......))))......))))))))..))).)).(((((....-.))-----)))..... ( -26.80, z-score = -2.01, R) >consensus GGAAAGCGAAGUGCGUUAAAUAAUAAGACGAAUUGCCGUCUUAGCAUGCUCAAAAUGUUUCGUCUUAAUGCAAUCUUGGCCGUCUCCUCC_AAA_____GACCACUU ...........((((((((....(((((((......)))))))((((.......))))......))))))))................................... (-10.75 = -10.57 + -0.19)

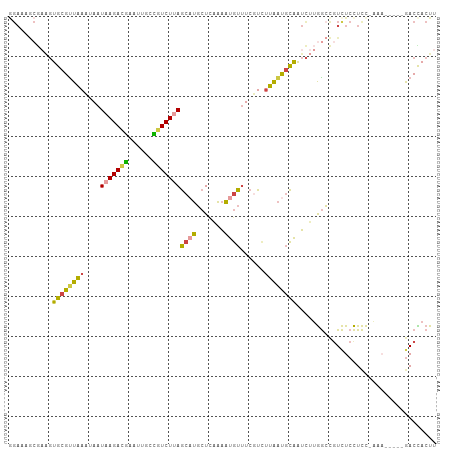
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:42:34 2011