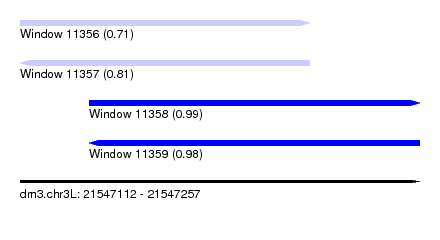
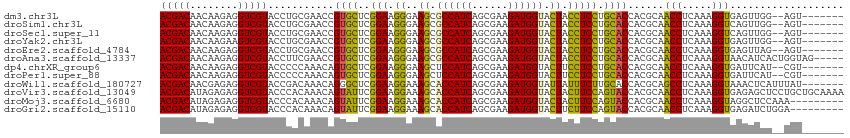
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 21,547,112 – 21,547,257 |
| Length | 145 |
| Max. P | 0.988952 |

| Location | 21,547,112 – 21,547,217 |
|---|---|
| Length | 105 |
| Sequences | 12 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 84.58 |
| Shannon entropy | 0.32683 |
| G+C content | 0.52814 |
| Mean single sequence MFE | -36.11 |
| Consensus MFE | -26.73 |
| Energy contribution | -26.00 |
| Covariance contribution | -0.73 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.74 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.710523 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
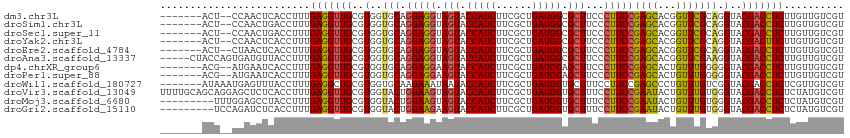
>dm3.chr3L 21547112 105 + 24543557 -------ACU--CCAACUCACCUUUGAGGUUGCGUGGUGCAGGAGGUAGUACCAUCUUCGCUGAUGGCGCUUCCCUUCCGAGCACGGUUCGCAGGUACGACCUCUUGUUGUCGU -------...--((..((((....))))..(((((.((((.(((((.((..(((((......)))))..))...)))))..)))).)..)))))).(((((........))))) ( -36.40, z-score = -1.06, R) >droSim1.chr3L 20883131 105 + 22553184 -------ACU--CCAACUGACCUUUGAGGUUGCGUGGUGCAGGAGGUAGUACCAUCUUCGCUGAUGGCGCUUCCCUUCCGAGCACGGUUCGCAGGUACGACCUCUUGUUGUCGU -------...--......(((....((((((((((.((((.(((((.((..(((((......)))))..))...)))))..)))).)..))))......))))).....))).. ( -36.20, z-score = -0.78, R) >droSec1.super_11 1467220 105 + 2888827 -------ACU--CCAACUGACCUUUGAGGUUGCGUGGUGCAGGAGGUAGUACCAUCUUCGCUGAUGGCGCUUCCCUUCCGAGCACGGUUCGCAGGUACGACCUCUUGUUGUCGU -------...--......(((....((((((((((.((((.(((((.((..(((((......)))))..))...)))))..)))).)..))))......))))).....))).. ( -36.20, z-score = -0.78, R) >droYak2.chr3L 5230222 105 - 24197627 -------ACU--CCAACUCACCUUUGAGGUUGCGUGGUGCAGGAGGUAGUACCAUCUUCGCUGAUGGCGCUUCCCUUCCGAGCACGGUUCGCAGGUACGACUUCUUGUUGUCGU -------...--((..((((....))))..(((((.((((.(((((.((..(((((......)))))..))...)))))..)))).)..)))))).(((((........))))) ( -37.60, z-score = -1.56, R) >droEre2.scaffold_4784 21201239 105 + 25762168 -------ACU--CUAACUCACCUUUGAGGUUGCGUGGUGCAGGAGGUAGUACCAUCUUCGCUGAUGGCGCUUCCCUUCCGAGCACGGUUCGCAGGUACGACCUCUUGUUGUCGU -------...--....((((....)))).((((((.((((.(((((.((..(((((......)))))..))...)))))..)))).)..)))))..(((((........))))) ( -35.70, z-score = -0.96, R) >droAna3.scaffold_13337 6203179 109 - 23293914 -----CUACCAGUGAUGUUACCUUUGAGGUUGCGUGGUGCAGGAGGUAGUACCAUCUUCGCUGAUGGCGCUUCCCUUCCGAGCACGGUUCGAAGGUACGACCUCUUGUUGUCGU -----..(((((..((.(((....))).))..).))))(((((((((((..(((((......)))))..))..(((((.((((...)))))))))....)))))))))...... ( -39.30, z-score = -1.67, R) >dp4.chrXR_group6 305066 105 - 13314419 -------ACG--AUGAAUCACCUUUGAGGUUGCGUGGUGCAGGAGGAAGUACCAUCUUCGCUGAUGGAGCUUCCCUUCCGAGCACUGUUUGGGGGUACGACCUCUUGUUGUCGU -------(((--(..(...(((.....)))...(..((((.(((((((((.(((((......))))).))))))..)))..))))..)..((((......))))...)..)))) ( -38.80, z-score = -1.66, R) >droPer1.super_88 176201 105 - 213009 -------ACG--AUGAAUCACCUUUGAGGUUGCGUGGUGCAGGAGGAAGUACCAUCUUCGCUGAUGGAGCUUCCCUUCCGAGCACUGUUUGGGGGUACGACCUCUUGUUGUCGU -------(((--(..(...(((.....)))...(..((((.(((((((((.(((((......))))).))))))..)))..))))..)..((((......))))...)..)))) ( -38.80, z-score = -1.66, R) >droWil1.scaffold_180727 1121509 107 + 2741493 -------AUAAAUGAGUUUACCUUUGAGGCUGCGUGGUGCAAGAAAUAAUACCAUCUUCGCUGAUGGUGCUUUCCUUCCGAGCCCUGUUUGUCGGUACGACCUCUCGUUGUCGU -------...((((((...........((((.((.((.((.(....)...((((((......))))))))...))...))))))......((((...))))..))))))..... ( -24.30, z-score = 0.09, R) >droVir3.scaffold_13049 13694935 114 + 25233164 UUUUGCAGCAGGAGCUCUCACCUUUGAGGUUGCGUGGUACUGGAAGUAGUACCAUCUUCGCUGAUGGUGCUUUCCUUCCGAAUACUGUUUGUGGGUACGACCUCUCUAUGUCGU ....((.((((.(((.((((....))))))).)((.((((((((((.(((((((((......)))))))))...)))))...(((.....)))))))).)).......))).)) ( -37.70, z-score = -1.83, R) >droMoj3.scaffold_6680 11112430 105 - 24764193 ---------UUUGGAGCCUACCUUUGAGGUUGCGUGGUACUGGAAGUAGUACCAUCUUCGCUGAUGGUGCUUUCCUUCCGAAUACUGUUUGUGGGUACGACCUCUCUAUGUCGU ---------..((((((..(((.....))).))((.((((((((((.(((((((((......)))))))))...)))))...(((.....)))))))).))...))))...... ( -35.70, z-score = -2.68, R) >droGri2.scaffold_15110 9023422 105 - 24565398 ---------UCCAGAUCUCACCUUUGAGGUUGCGUGGUACUGGAAGAAGUACCAUCUUCGCUGAUGGUGCUUUCCUUCCGAAUACUGUUUGUGGGUACGACCUCUCUAUGUCGU ---------(((((((((((....)))))))..(..(((.((((((((((((((((......))))))))))..))))))..)))..)...)))).(((((........))))) ( -36.60, z-score = -3.04, R) >consensus _______ACU__CGAACUCACCUUUGAGGUUGCGUGGUGCAGGAGGUAGUACCAUCUUCGCUGAUGGCGCUUCCCUUCCGAGCACGGUUCGCAGGUACGACCUCUUGUUGUCGU .........................(((((((..(..(((.(((((.(((.(((((......))))).)))...)))))((((...))))))).)..))))))).......... (-26.73 = -26.00 + -0.73)
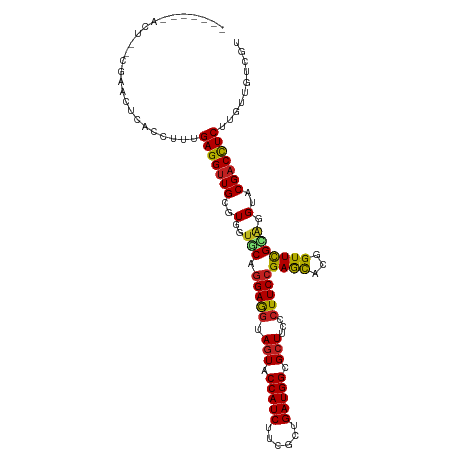
| Location | 21,547,112 – 21,547,217 |
|---|---|
| Length | 105 |
| Sequences | 12 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 84.58 |
| Shannon entropy | 0.32683 |
| G+C content | 0.52814 |
| Mean single sequence MFE | -31.96 |
| Consensus MFE | -24.40 |
| Energy contribution | -23.40 |
| Covariance contribution | -1.00 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.76 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.813161 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 21547112 105 - 24543557 ACGACAACAAGAGGUCGUACCUGCGAACCGUGCUCGGAAGGGAAGCGCCAUCAGCGAAGAUGGUACUACCUCCUGCACCACGCAACCUCAAAGGUGAGUUGG--AGU------- .((((.((..(((((......((((....((((..(((.((..((..(((((......)))))..)).))))).))))..)))))))))....))..)))).--...------- ( -35.20, z-score = -1.29, R) >droSim1.chr3L 20883131 105 - 22553184 ACGACAACAAGAGGUCGUACCUGCGAACCGUGCUCGGAAGGGAAGCGCCAUCAGCGAAGAUGGUACUACCUCCUGCACCACGCAACCUCAAAGGUCAGUUGG--AGU------- ..(((.....(((((......((((....((((..(((.((..((..(((((......)))))..)).))))).))))..)))))))))....)))......--...------- ( -34.80, z-score = -1.26, R) >droSec1.super_11 1467220 105 - 2888827 ACGACAACAAGAGGUCGUACCUGCGAACCGUGCUCGGAAGGGAAGCGCCAUCAGCGAAGAUGGUACUACCUCCUGCACCACGCAACCUCAAAGGUCAGUUGG--AGU------- ..(((.....(((((......((((....((((..(((.((..((..(((((......)))))..)).))))).))))..)))))))))....)))......--...------- ( -34.80, z-score = -1.26, R) >droYak2.chr3L 5230222 105 + 24197627 ACGACAACAAGAAGUCGUACCUGCGAACCGUGCUCGGAAGGGAAGCGCCAUCAGCGAAGAUGGUACUACCUCCUGCACCACGCAACCUCAAAGGUGAGUUGG--AGU------- (((((........))))).((((((....((((..(((.((..((..(((((......)))))..)).))))).))))..))))..((((....))))..))--...------- ( -34.80, z-score = -1.53, R) >droEre2.scaffold_4784 21201239 105 - 25762168 ACGACAACAAGAGGUCGUACCUGCGAACCGUGCUCGGAAGGGAAGCGCCAUCAGCGAAGAUGGUACUACCUCCUGCACCACGCAACCUCAAAGGUGAGUUAG--AGU------- (((((........)))))...((((....((((..(((.((..((..(((((......)))))..)).))))).))))..))))..((((....))))....--...------- ( -33.20, z-score = -1.13, R) >droAna3.scaffold_13337 6203179 109 + 23293914 ACGACAACAAGAGGUCGUACCUUCGAACCGUGCUCGGAAGGGAAGCGCCAUCAGCGAAGAUGGUACUACCUCCUGCACCACGCAACCUCAAAGGUAACAUCACUGGUAG----- (((((........)))))........(((((((..(((.((..((..(((((......)))))..)).))))).))))......(((.....))).........)))..----- ( -32.00, z-score = -1.19, R) >dp4.chrXR_group6 305066 105 + 13314419 ACGACAACAAGAGGUCGUACCCCCAAACAGUGCUCGGAAGGGAAGCUCCAUCAGCGAAGAUGGUACUUCCUCCUGCACCACGCAACCUCAAAGGUGAUUCAU--CGU------- ..........(((((.((........)).((((..(((..(((((..(((((......)))))..)))))))).))))......)))))...((((...)))--)..------- ( -30.40, z-score = -1.33, R) >droPer1.super_88 176201 105 + 213009 ACGACAACAAGAGGUCGUACCCCCAAACAGUGCUCGGAAGGGAAGCUCCAUCAGCGAAGAUGGUACUUCCUCCUGCACCACGCAACCUCAAAGGUGAUUCAU--CGU------- ..........(((((.((........)).((((..(((..(((((..(((((......)))))..)))))))).))))......)))))...((((...)))--)..------- ( -30.40, z-score = -1.33, R) >droWil1.scaffold_180727 1121509 107 - 2741493 ACGACAACGAGAGGUCGUACCGACAAACAGGGCUCGGAAGGAAAGCACCAUCAGCGAAGAUGGUAUUAUUUCUUGCACCACGCAGCCUCAAAGGUAAACUCAUUUAU------- ..........(((....((((........(((((((..((((((..((((((......))))))....))))))......)).)))))....))))..)))......------- ( -27.30, z-score = -1.74, R) >droVir3.scaffold_13049 13694935 114 - 25233164 ACGACAUAGAGAGGUCGUACCCACAAACAGUAUUCGGAAGGAAAGCACCAUCAGCGAAGAUGGUACUACUUCCAGUACCACGCAACCUCAAAGGUGAGAGCUCCUGCUGCAAAA (((((........)))))...........(((((.(((((...((.((((((......)))))).)).))))))))))...((...((((....))))(((....))))).... ( -32.30, z-score = -2.15, R) >droMoj3.scaffold_6680 11112430 105 + 24764193 ACGACAUAGAGAGGUCGUACCCACAAACAGUAUUCGGAAGGAAAGCACCAUCAGCGAAGAUGGUACUACUUCCAGUACCACGCAACCUCAAAGGUAGGCUCCAAA--------- (((((........)))))...........(((((.(((((...((.((((((......)))))).)).))))))))))...((.(((.....)))..))......--------- ( -28.70, z-score = -2.59, R) >droGri2.scaffold_15110 9023422 105 + 24565398 ACGACAUAGAGAGGUCGUACCCACAAACAGUAUUCGGAAGGAAAGCACCAUCAGCGAAGAUGGUACUUCUUCCAGUACCACGCAACCUCAAAGGUGAGAUCUGGA--------- (((((........)))))..(((......(((((.((((((..((.((((((......)))))).)))))))))))))........((((....))))...))).--------- ( -29.60, z-score = -1.91, R) >consensus ACGACAACAAGAGGUCGUACCUGCAAACAGUGCUCGGAAGGGAAGCGCCAUCAGCGAAGAUGGUACUACCUCCUGCACCACGCAACCUCAAAGGUGAGUUCG__AGU_______ (((((........)))))...........((((..(((.((..((.((((((......)))))).)).))))).))))......(((.....)))................... (-24.40 = -23.40 + -1.00)
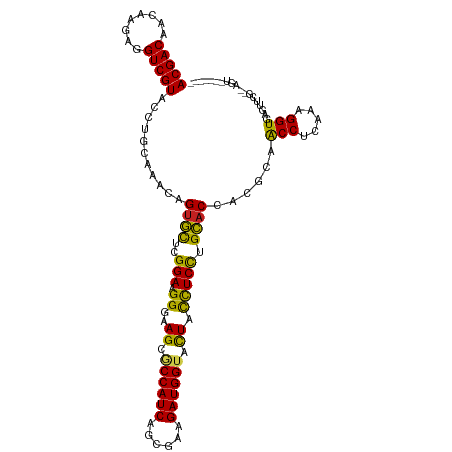
| Location | 21,547,137 – 21,547,257 |
|---|---|
| Length | 120 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.16 |
| Shannon entropy | 0.20391 |
| G+C content | 0.48264 |
| Mean single sequence MFE | -41.79 |
| Consensus MFE | -33.32 |
| Energy contribution | -32.49 |
| Covariance contribution | -0.83 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.92 |
| Structure conservation index | 0.80 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.34 |
| SVM RNA-class probability | 0.988952 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 21547137 120 + 24543557 UGGUGCAGGAGGUAGUACCAUCUUCGCUGAUGGCGCUUCCCUUCCGAGCACGGUUCGCAGGUACGACCUCUUGUUGUCGUACGAAGUCGCAGAGGGUUUGUCCAAUUUUGCAAGAUUUUU ..((((.(((((.((..(((((......)))))..))...)))))..)))).........(((((((........)))))))(((((((((((.((.....))...)))))..)))))). ( -41.20, z-score = -1.65, R) >droSim1.chr3L 20883156 120 + 22553184 UGGUGCAGGAGGUAGUACCAUCUUCGCUGAUGGCGCUUCCCUUCCGAGCACGGUUCGCAGGUACGACCUCUUGUUGUCGUACGAAGUCGCAGAGGGUUUGUCCAAUUUUGCAAGAUUUUU ..((((.(((((.((..(((((......)))))..))...)))))..)))).........(((((((........)))))))(((((((((((.((.....))...)))))..)))))). ( -41.20, z-score = -1.65, R) >droSec1.super_11 1467245 120 + 2888827 UGGUGCAGGAGGUAGUACCAUCUUCGCUGAUGGCGCUUCCCUUCCGAGCACGGUUCGCAGGUACGACCUCUUGUUGUCGUACGAAGUCGCAGAGGGUUUGUCCAAUUUUGCAAGAUUUUU ..((((.(((((.((..(((((......)))))..))...)))))..)))).........(((((((........)))))))(((((((((((.((.....))...)))))..)))))). ( -41.20, z-score = -1.65, R) >droYak2.chr3L 5230247 120 - 24197627 UGGUGCAGGAGGUAGUACCAUCUUCGCUGAUGGCGCUUCCCUUCCGAGCACGGUUCGCAGGUACGACUUCUUGUUGUCGUACAAAGUCGCAGAGGGUUUGUCCAAUUUUGCAAGAUUUUC ..((((.(((((.((..(((((......)))))..))...)))))..)))).........(((((((........)))))))(((((((((((.((.....))...)))))..)))))). ( -42.00, z-score = -2.38, R) >droEre2.scaffold_4784 21201264 120 + 25762168 UGGUGCAGGAGGUAGUACCAUCUUCGCUGAUGGCGCUUCCCUUCCGAGCACGGUUCGCAGGUACGACCUCUUGUUGUCGUACGAAAUCGCAGAGGGUUUGUCCAAUUUUGCAAGAUUUUC ..((((.(((((.((..(((((......)))))..))...)))))..)))).........(((((((........)))))))(((((((((((.((.....))...)))))..)))))). ( -41.20, z-score = -1.83, R) >droAna3.scaffold_13337 6203208 120 - 23293914 UGGUGCAGGAGGUAGUACCAUCUUCGCUGAUGGCGCUUCCCUUCCGAGCACGGUUCGAAGGUACGACCUCUUGUUGUCGUACAAAGUCGCAAAGGGUUUGUCCAAUUUUGCAGGAUUUUC ..((((.(((((.((..(((((......)))))..))...)))))..))))(((((((..(((((((........)))))))....))(((((.((.....))...))))).)))))... ( -42.50, z-score = -2.41, R) >dp4.chrXR_group6 305091 120 - 13314419 UGGUGCAGGAGGAAGUACCAUCUUCGCUGAUGGAGCUUCCCUUCCGAGCACUGUUUGGGGGUACGACCUCUUGUUGUCGUACAAAGUCACAGAGGGUUUGUCCAAUUUUGUAUGAUUUUC .(((((.(((((((((.(((((......))))).))))))..)))..)))))....((((......)))).....((((((((((((.(((((...)))))...)))))))))))).... ( -44.00, z-score = -2.80, R) >droPer1.super_88 176226 120 - 213009 UGGUGCAGGAGGAAGUACCAUCUUCGCUGAUGGAGCUUCCCUUCCGAGCACUGUUUGGGGGUACGACCUCUUGUUGUCGUACAAAGUCACAGAGGGUUUGUCCAAUUUUGUAUGAUUUUC .(((((.(((((((((.(((((......))))).))))))..)))..)))))....((((......)))).....((((((((((((.(((((...)))))...)))))))))))).... ( -44.00, z-score = -2.80, R) >droWil1.scaffold_180727 1121536 120 + 2741493 UGGUGCAAGAAAUAAUACCAUCUUCGCUGAUGGUGCUUUCCUUCCGAGCCCUGUUUGUCGGUACGACCUCUCGUUGUCGUACAAAGUCGCAAAGGGUUUGUCCAAUUUUGCAAACUUUUU ...(((((((.....(((((((......))))))).........((((((((...((.(.(((((((........)))))))...).))...)))))))).....)))))))........ ( -38.50, z-score = -3.69, R) >droVir3.scaffold_13049 13694969 120 + 25233164 UGGUACUGGAAGUAGUACCAUCUUCGCUGAUGGUGCUUUCCUUCCGAAUACUGUUUGUGGGUACGACCUCUCUAUGUCGUACAAAGUCGCAAAGGGUUUGUCCAAUUUUGUAAAAUUUUC (((.((((((((.(((((((((......)))))))))...))))))...(((.((((((((((((((........)))))))....))))))).)))..)))))................ ( -41.80, z-score = -4.92, R) >droMoj3.scaffold_6680 11112455 120 - 24764193 UGGUACUGGAAGUAGUACCAUCUUCGCUGAUGGUGCUUUCCUUCCGAAUACUGUUUGUGGGUACGACCUCUCUAUGUCGUACAAAGUCGCAAAGGGUUUGUCCAAUUUUGCAAUAUUUUC (((.((((((((.(((((((((......)))))))))...))))))...(((.((((((((((((((........)))))))....))))))).)))..)))))................ ( -41.80, z-score = -4.62, R) >droGri2.scaffold_15110 9023447 120 - 24565398 UGGUACUGGAAGAAGUACCAUCUUCGCUGAUGGUGCUUUCCUUCCGAAUACUGUUUGUGGGUACGACCUCUCUAUGUCGUACAAAGUCGCAAAGGGUUUGUCCAAUUUUGCAAUAUUUUC (((.((((((((((((((((((......))))))))))..))))))...(((.((((((((((((((........)))))))....))))))).)))..)))))................ ( -42.10, z-score = -4.62, R) >consensus UGGUGCAGGAGGUAGUACCAUCUUCGCUGAUGGCGCUUCCCUUCCGAGCACGGUUCGCAGGUACGACCUCUUGUUGUCGUACAAAGUCGCAGAGGGUUUGUCCAAUUUUGCAAGAUUUUC ..(((..(((((.(((.(((((......))))).)))...)))))...))).........(((((((........)))))))......(((((.((.....))...)))))......... (-33.32 = -32.49 + -0.83)
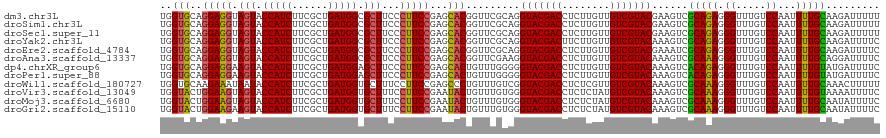
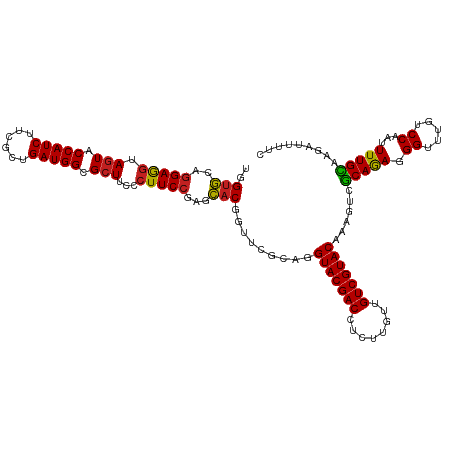
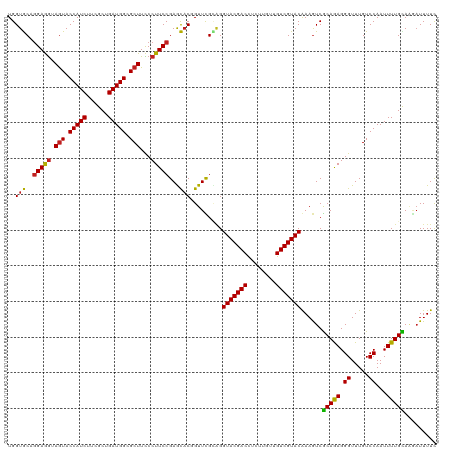
| Location | 21,547,137 – 21,547,257 |
|---|---|
| Length | 120 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.16 |
| Shannon entropy | 0.20391 |
| G+C content | 0.48264 |
| Mean single sequence MFE | -35.46 |
| Consensus MFE | -29.11 |
| Energy contribution | -28.02 |
| Covariance contribution | -1.09 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.54 |
| Structure conservation index | 0.82 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.94 |
| SVM RNA-class probability | 0.975735 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 21547137 120 - 24543557 AAAAAUCUUGCAAAAUUGGACAAACCCUCUGCGACUUCGUACGACAACAAGAGGUCGUACCUGCGAACCGUGCUCGGAAGGGAAGCGCCAUCAGCGAAGAUGGUACUACCUCCUGCACCA .......(((((.....((......))..(((((((((............)))))))))..)))))...((((..(((.((..((..(((((......)))))..)).))))).)))).. ( -37.80, z-score = -2.40, R) >droSim1.chr3L 20883156 120 - 22553184 AAAAAUCUUGCAAAAUUGGACAAACCCUCUGCGACUUCGUACGACAACAAGAGGUCGUACCUGCGAACCGUGCUCGGAAGGGAAGCGCCAUCAGCGAAGAUGGUACUACCUCCUGCACCA .......(((((.....((......))..(((((((((............)))))))))..)))))...((((..(((.((..((..(((((......)))))..)).))))).)))).. ( -37.80, z-score = -2.40, R) >droSec1.super_11 1467245 120 - 2888827 AAAAAUCUUGCAAAAUUGGACAAACCCUCUGCGACUUCGUACGACAACAAGAGGUCGUACCUGCGAACCGUGCUCGGAAGGGAAGCGCCAUCAGCGAAGAUGGUACUACCUCCUGCACCA .......(((((.....((......))..(((((((((............)))))))))..)))))...((((..(((.((..((..(((((......)))))..)).))))).)))).. ( -37.80, z-score = -2.40, R) >droYak2.chr3L 5230247 120 + 24197627 GAAAAUCUUGCAAAAUUGGACAAACCCUCUGCGACUUUGUACGACAACAAGAAGUCGUACCUGCGAACCGUGCUCGGAAGGGAAGCGCCAUCAGCGAAGAUGGUACUACCUCCUGCACCA .......(((((.....((......))...........(((((((........))))))).)))))...((((..(((.((..((..(((((......)))))..)).))))).)))).. ( -36.00, z-score = -2.36, R) >droEre2.scaffold_4784 21201264 120 - 25762168 GAAAAUCUUGCAAAAUUGGACAAACCCUCUGCGAUUUCGUACGACAACAAGAGGUCGUACCUGCGAACCGUGCUCGGAAGGGAAGCGCCAUCAGCGAAGAUGGUACUACCUCCUGCACCA ((((...(((((.....((......))..)))))))))(((((((........))))))).........((((..(((.((..((..(((((......)))))..)).))))).)))).. ( -36.80, z-score = -2.05, R) >droAna3.scaffold_13337 6203208 120 + 23293914 GAAAAUCCUGCAAAAUUGGACAAACCCUUUGCGACUUUGUACGACAACAAGAGGUCGUACCUUCGAACCGUGCUCGGAAGGGAAGCGCCAUCAGCGAAGAUGGUACUACCUCCUGCACCA (((.....((((((...((......)))))))).....(((((((........))))))).))).....((((..(((.((..((..(((((......)))))..)).))))).)))).. ( -36.50, z-score = -2.30, R) >dp4.chrXR_group6 305091 120 + 13314419 GAAAAUCAUACAAAAUUGGACAAACCCUCUGUGACUUUGUACGACAACAAGAGGUCGUACCCCCAAACAGUGCUCGGAAGGGAAGCUCCAUCAGCGAAGAUGGUACUUCCUCCUGCACCA ...............(((((((.......)))......(((((((........)))))))..))))...((((..(((..(((((..(((((......)))))..)))))))).)))).. ( -35.40, z-score = -2.50, R) >droPer1.super_88 176226 120 + 213009 GAAAAUCAUACAAAAUUGGACAAACCCUCUGUGACUUUGUACGACAACAAGAGGUCGUACCCCCAAACAGUGCUCGGAAGGGAAGCUCCAUCAGCGAAGAUGGUACUUCCUCCUGCACCA ...............(((((((.......)))......(((((((........)))))))..))))...((((..(((..(((((..(((((......)))))..)))))))).)))).. ( -35.40, z-score = -2.50, R) >droWil1.scaffold_180727 1121536 120 - 2741493 AAAAAGUUUGCAAAAUUGGACAAACCCUUUGCGACUUUGUACGACAACGAGAGGUCGUACCGACAAACAGGGCUCGGAAGGAAAGCACCAUCAGCGAAGAUGGUAUUAUUUCUUGCACCA ..((((((.(((((...((......)))))))))))))(((((((........))))))).........((((..(((((..((..((((((......)))))).)).))))).)).)). ( -32.90, z-score = -1.81, R) >droVir3.scaffold_13049 13694969 120 - 25233164 GAAAAUUUUACAAAAUUGGACAAACCCUUUGCGACUUUGUACGACAUAGAGAGGUCGUACCCACAAACAGUAUUCGGAAGGAAAGCACCAUCAGCGAAGAUGGUACUACUUCCAGUACCA ................(((.(((.....))).......(((((((........))))))))))......(((((.(((((...((.((((((......)))))).)).)))))))))).. ( -29.70, z-score = -2.42, R) >droMoj3.scaffold_6680 11112455 120 + 24764193 GAAAAUAUUGCAAAAUUGGACAAACCCUUUGCGACUUUGUACGACAUAGAGAGGUCGUACCCACAAACAGUAUUCGGAAGGAAAGCACCAUCAGCGAAGAUGGUACUACUUCCAGUACCA .......(((((((...((......)))))))))....(((((((........))))))).........(((((.(((((...((.((((((......)))))).)).)))))))))).. ( -34.70, z-score = -3.78, R) >droGri2.scaffold_15110 9023447 120 + 24565398 GAAAAUAUUGCAAAAUUGGACAAACCCUUUGCGACUUUGUACGACAUAGAGAGGUCGUACCCACAAACAGUAUUCGGAAGGAAAGCACCAUCAGCGAAGAUGGUACUUCUUCCAGUACCA .......(((((((...((......)))))))))....(((((((........))))))).........(((((.((((((..((.((((((......)))))).))))))))))))).. ( -34.70, z-score = -3.55, R) >consensus GAAAAUCUUGCAAAAUUGGACAAACCCUCUGCGACUUUGUACGACAACAAGAGGUCGUACCUGCAAACAGUGCUCGGAAGGGAAGCGCCAUCAGCGAAGAUGGUACUACCUCCUGCACCA ........((((.....((......))..)))).....(((((((........))))))).........((((..(((.((..((.((((((......)))))).)).))))).)))).. (-29.11 = -28.02 + -1.09)

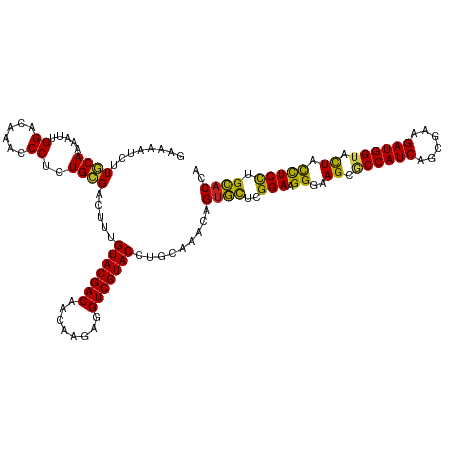
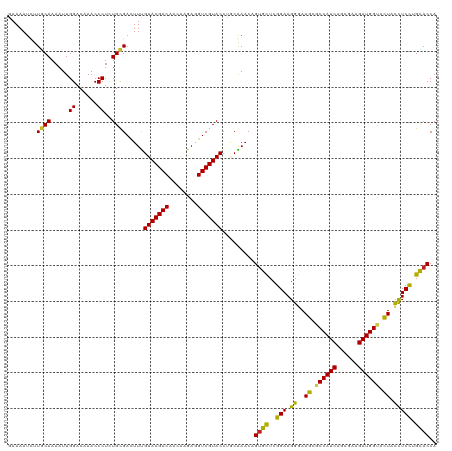
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:42:25 2011